Application of circ-0000423 to preparation or screening of colon cancer drugs
A colon cancer, colorectal cancer technology, applied in the field of biomedicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
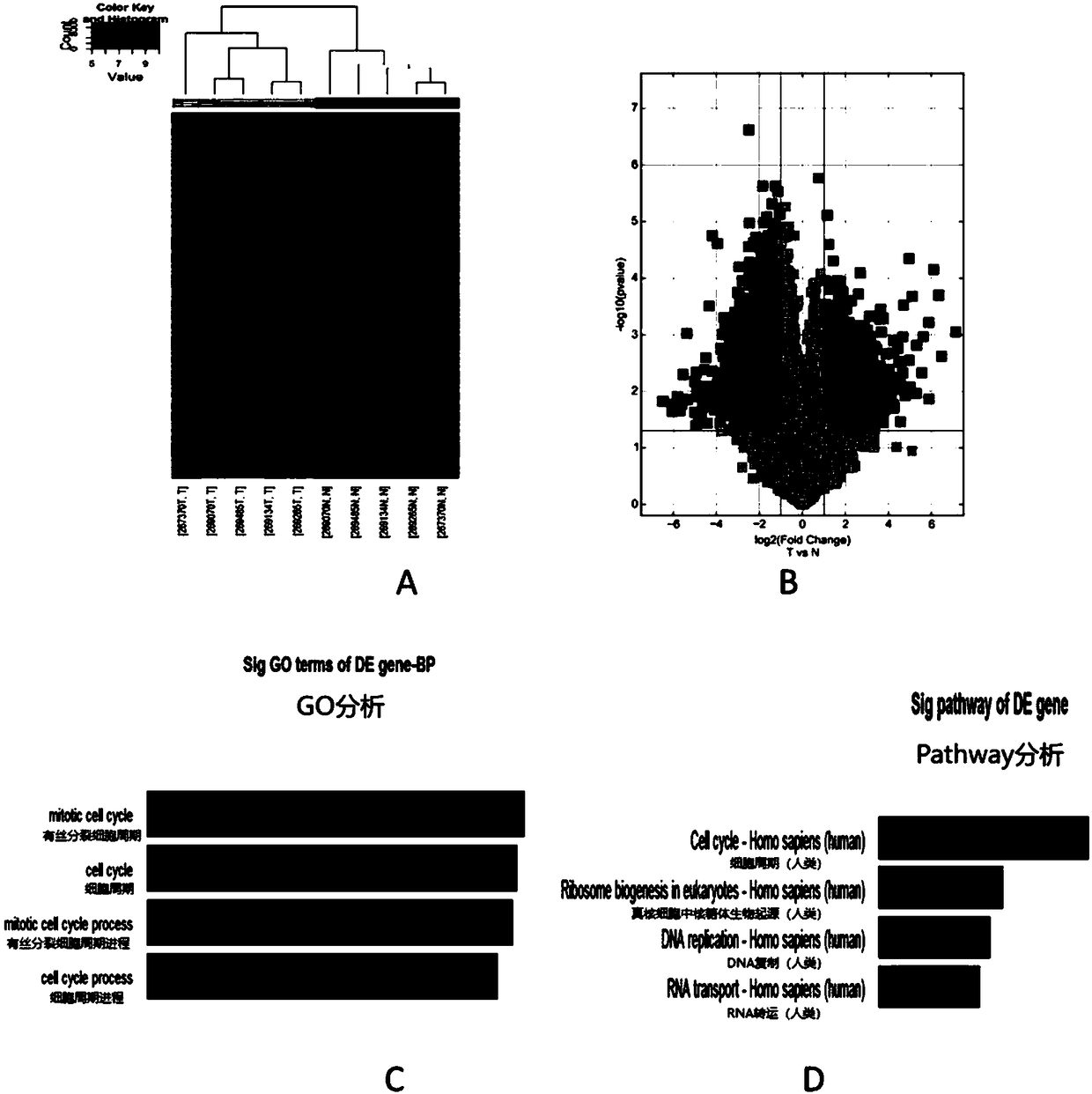
[0030] (1) The gene chip was completed by Beijing Kangcheng Company; the human circRNAs / mRNA 4×180K Microarray co-expression chip V.2.0 platform customized by Agilent of the United States was used, covering 4425 circRNAs and 32197 mRNAs; for the original data, Lowess (locally weighted scatterplot smoothing) and other methods to normalize the signal value, then carry out differential gene screening. SAM (Significance Analysis of Microarrays) software was used to analyze the differential genes, and the FDR was controlled within 5%. Using methods such as multiple method and multiple hypothesis testing, the circRNAs and mRNAs with differential expression under two or more conditions were calculated and screened respectively.
[0031] (2) GO functional annotation Gene ontology (Gene ontology, GO) functional annotation is one of the most widely used methods for gene functional annotation. The GO database (www.geneontology.org / ) was created by the Gene Ontology consortium. The GO da...
Embodiment 2
[0036] According to the tissue microarray, 4 transcripts with expression differences of more than 5 times, high abundance and good uniformity were further screened out of 41 circRNAs, of which 3 were up-regulated and 1 was down-regulated (Table 1) ( image 3 A-D).
[0037] Table 1. According to the chip results, 3 up-regulated and 1 down-regulated circRNAs were repeatedly screened
[0038]
[0039] Subsequently, we performed tissue PCR verification on the above four differentially expressed circRNAs in 20 pairs of colorectal cancers:
[0040] Extract total RNA from colon cancer tissue, synthesize cDNA according to the instructions of the TAKARA036A kit, and perform fluorescent quantitative PCR reaction (see Table 2 for primers): according to the principle of experiment repetition and control, set up three replicate wells for each group of samples each time, and set up a negative control compared with blank. Take the 10 μL system as an example according to the instructions:...
Embodiment 3
[0047] The expression of circ-0000423 in colon cancer cell lines HT29 and HCT116 was verified by designing specific back-to-back primers for circ-0000423 junction and performing RT-PCR (see Table 2 for primers of circ-000423). At the same time, in view of the special structure of circRNA and its insensitivity to RNase, RT-PCR successfully identified the existence of circ-0000423 in colorectal cancer cell lines after RNase R treatment ( Figure 4 ); at the same time, searching UCSC shows that: circ-0000423 is located on the PPP1R12A gene of the long arm of chromosome 12, formed by the circularization of exons 1 and 2 of PPP1R12A, with a length of 1138bp, and the sequence is shown in SEQ ID NO.1 ( Figure 5 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com