Method for eliminating three-dimensional genomics technology noise by utilizing exonuclease combination
A genomics and exonuclease technology, applied in special data processing applications, instruments, electronic digital data processing, etc., can solve the problems of increasing technical efficiency and result specificity, so as to increase technical efficiency and result specificity, and improve production Out of proportion, the effect of simplification and elimination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1. Evaluation of the combined effect of exonucleases and their application in eliminating linear plasmid DNA.
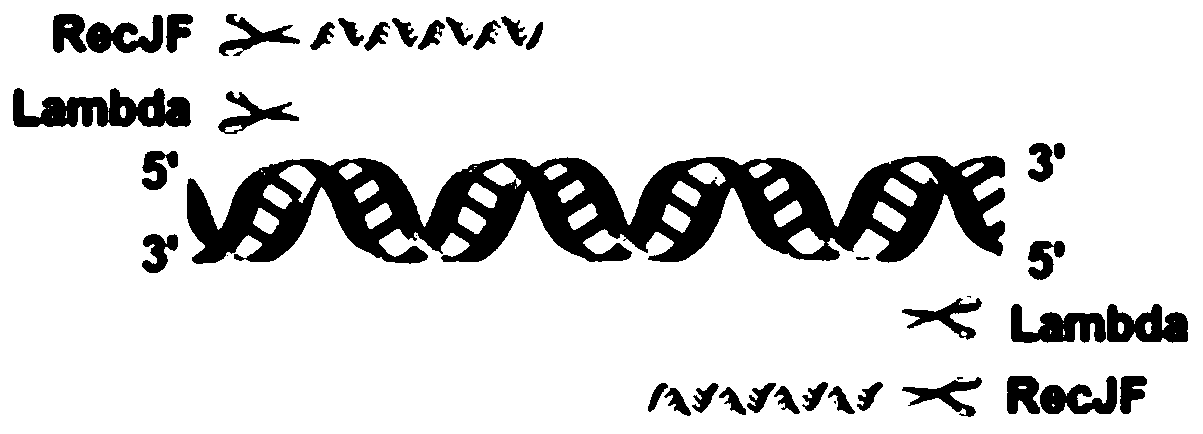
[0029] 1. Selection of exonuclease and exonuclease combination
[0030] The following exonucleases were selected from NEB (New England Biolabs), as shown in Table 1.
[0031] Table 1 Exonucleases and their characteristics related to linear removal experiments
[0032]
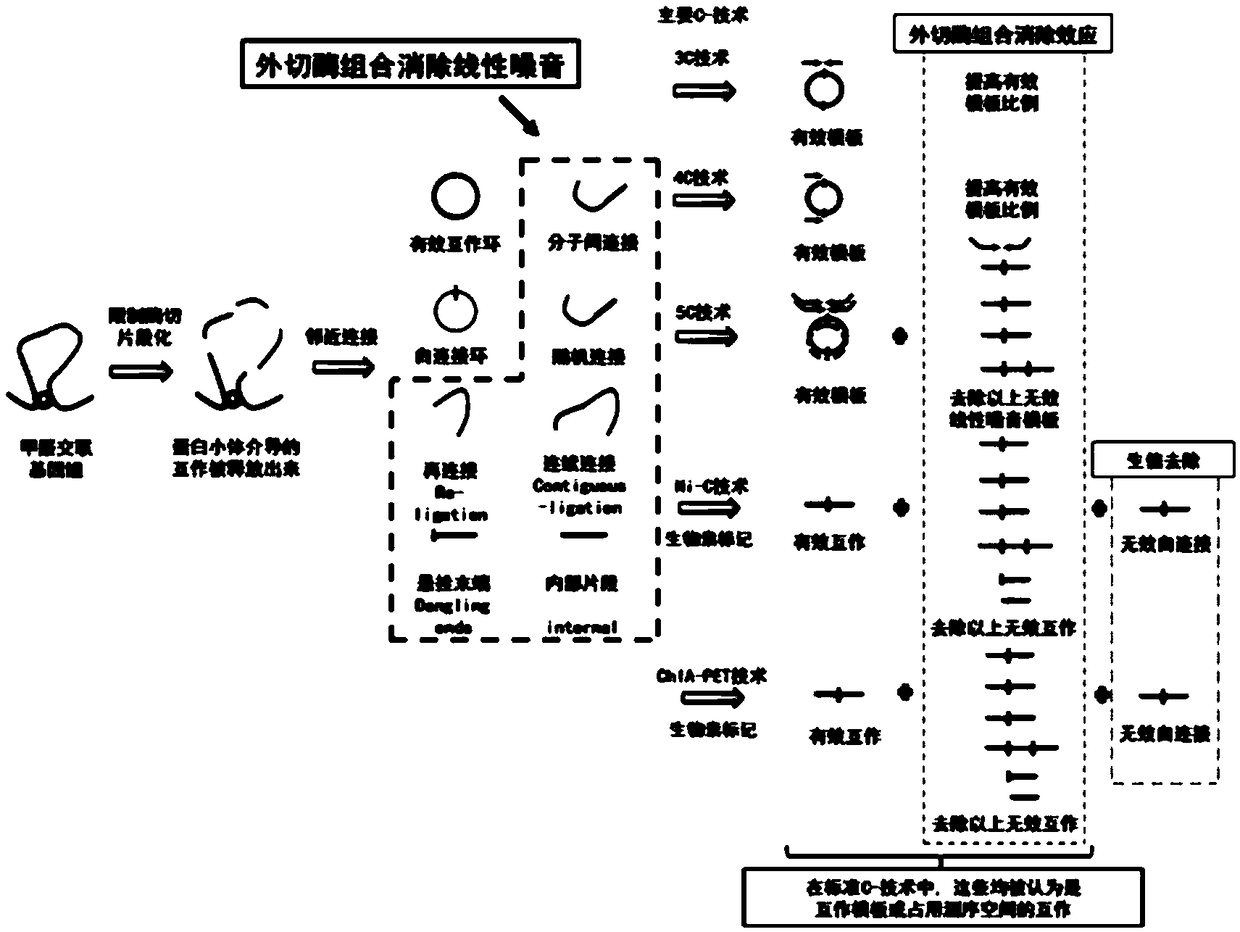
[0033]A single exonuclease has a single function. Combining two exonucleases with complementary functions will have more comprehensive functions and better removal effects than a single exonuclease. According to functional complementarity and related literature reports, Lambda and RecJF exonuclease combination (see image 3 ): LRL combination, Lambda and RecJF exonuclease combined and made to react in Lambdabuffer; LRC combination, Lambda and RecJF exonuclease combined and made to react in CutSmart buffer.
[0034] 2. Exonuclease Combination Elimination of Linear Plasmid DNA Evalu...
Embodiment 2
[0041] Example 2. Application of exonuclease combination in noise elimination in key steps of C-technology.
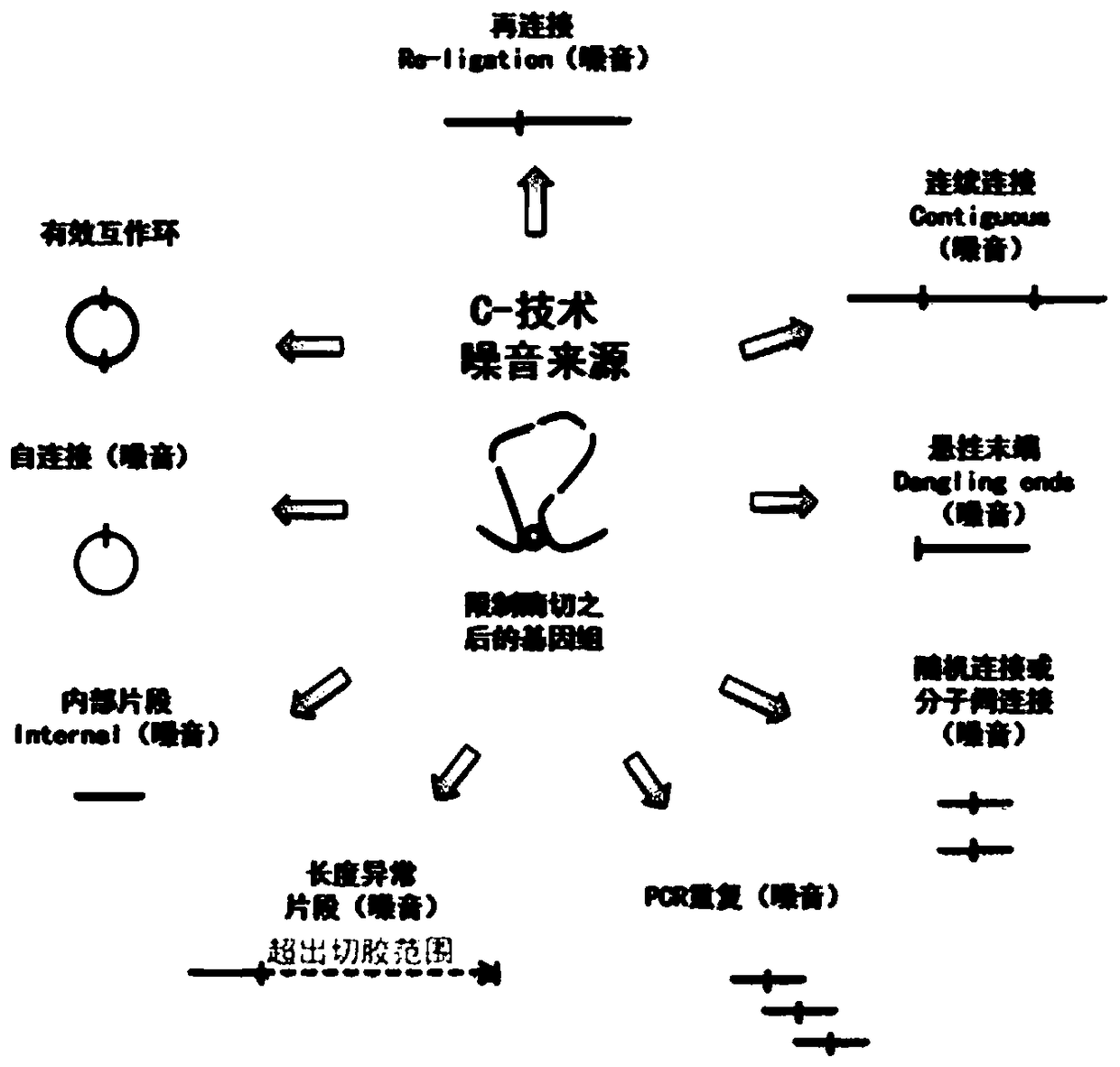
[0042] C-techniques are used for three-dimensional genomics chromatin conformation grasping, which is a critical step derived from proximity junctions. How to increase the ratio of circularized junction molecules is very important for the subsequent detection of DNA ligation efficiency. C-technique prior to the adjacent connection step, the major steps are identical (see figure 2 ). In this example, the combination of exonucleases verified in the plasmid test in Example 1 was used to eliminate the linear "noise" DNA existing after the adjacent connection in the C-technology, and its efficient elimination effect was successfully verified. In the current C-technology, the starting amount of cells used is from 0.01 to 25 million. In the experiment, it was found that about 300 ng of DNA library can be obtained from 0.1 million starting cells, and about 30 ng of DNA libra...
Embodiment 3
[0049] Example 3. Application of Exonuclease Combination in Hi-C Technology for Noise Elimination.
[0050] Hi-C is a high-throughput chromatin conformation capture technology based on 3C technology, which can capture the entire genome interaction mediated by proteosomes. Proximity connectivity is a key step in Hi-C technology. Proximity connections are currently noisy, and this step is one of the main reasons for the low effective interaction rate of standard Hi-C (see figure 1 and figure 2 ). Example 1 The exonuclease combination elimination method has been verified in the plasmid test. Example 2 The combined elimination method of exonucleases has been validated for noise reduction after the common core step of C-technique—“adjacent joining”. This example intends to demonstrate the elimination effect of the exonuclease combination noise elimination method applied in one of the specific C-technical members—Hi-C technology: increase the effective interaction ratio.
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com