Primer pair for assaying expression of CG5210 gene of tephritidae
A technology of CG5210 and gene expression, which is applied in the field of primer pairs for detection of CG5210 gene expression in fruit flies, can solve problems such as fungal infection, fruit rot and fall off, and achieve the effects of low cost, simple operation, and controlled diffusion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1. Preparation of primer pairs for detection of fruit fly CG5210 and Idgf4 gene expression
[0040] This embodiment provides a pair of primers for detecting the expression levels of CG5210 and Idgf4 genes in fruit flies, which are respectively denoted as CG5210-rt and Idgf4-rt. CG5210-rt is composed of two single-stranded DNAs shown in sequence 1 and sequence 2 in the sequence listing, the molar ratio of the two single-stranded DNAs in the primer pair is 1:1, and the two single-stranded DNAs are packaged independently. Idgf4-rt is composed of two single-stranded DNAs shown in sequence 3 and sequence 4 in the sequence listing, the molar ratio of the two single-stranded DNAs in the primer pair is 1:1, and the two single-stranded DNAs are packaged independently.
Embodiment 2
[0041] Example 2, using the primer pair of Example 1 to detect the expression of CG5210 and Idgf4 genes in fruit flies
[0042] Fruit flies to be tested: Guava fruit flies collected from Yunnan and Bactrocera dorsalis collected from Guangzhou, 50 first-instar larvae, 30 second-instar larvae, and third-instar early larvae (5 days after egg hatching) 30 instar larvae), 20 late third-instar larvae (7-day-old larvae after egg hatching), 6 early-stage pupae (1-day-old pupae since pupation), 6 mid-stage pupae (5-day-old pupae since pupation) 6, 6 late stage pupae (9 days old pupae since pupation), 10 immature adults after eclosion, 10 sexually mature adults 10 days old.
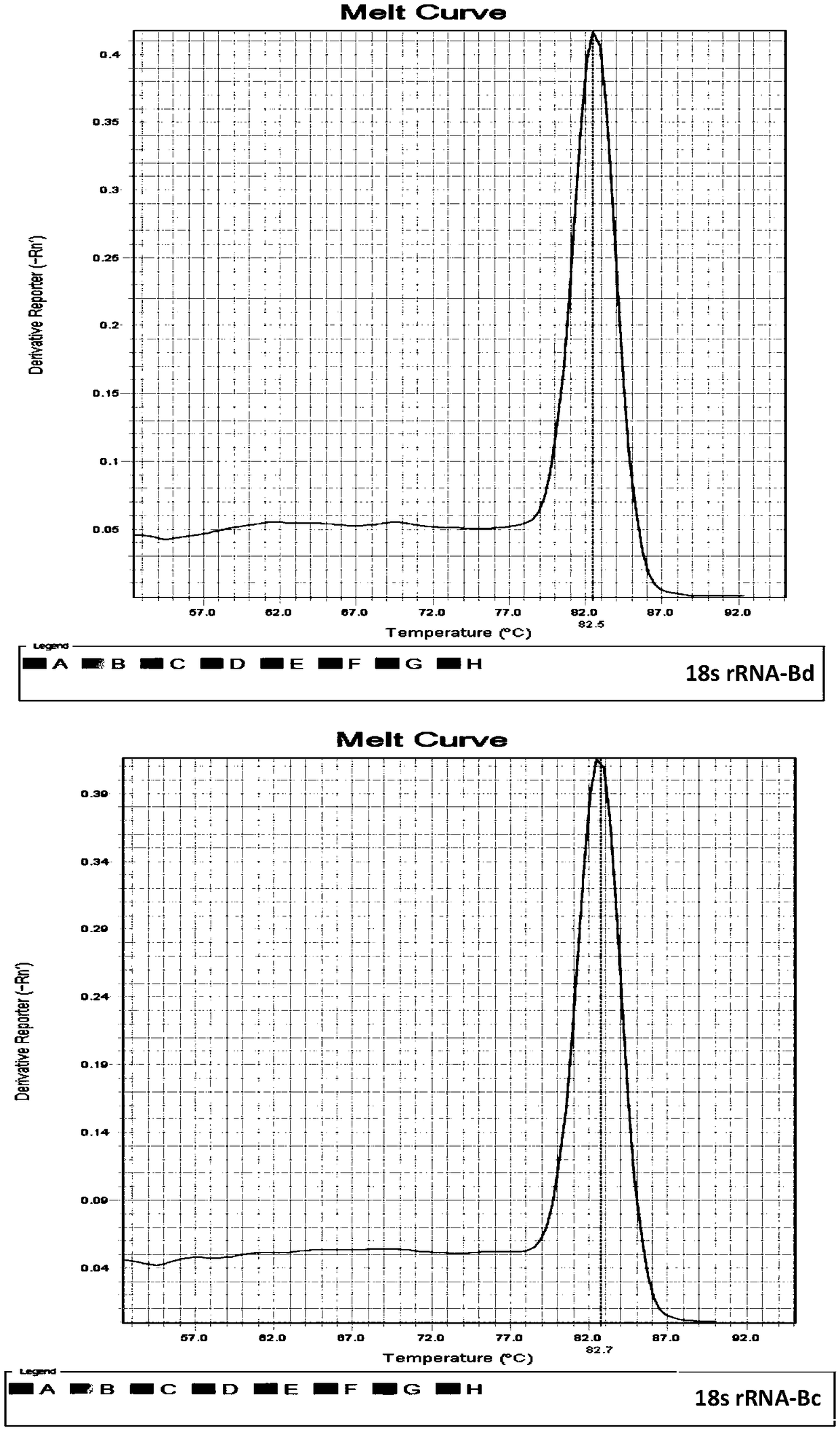
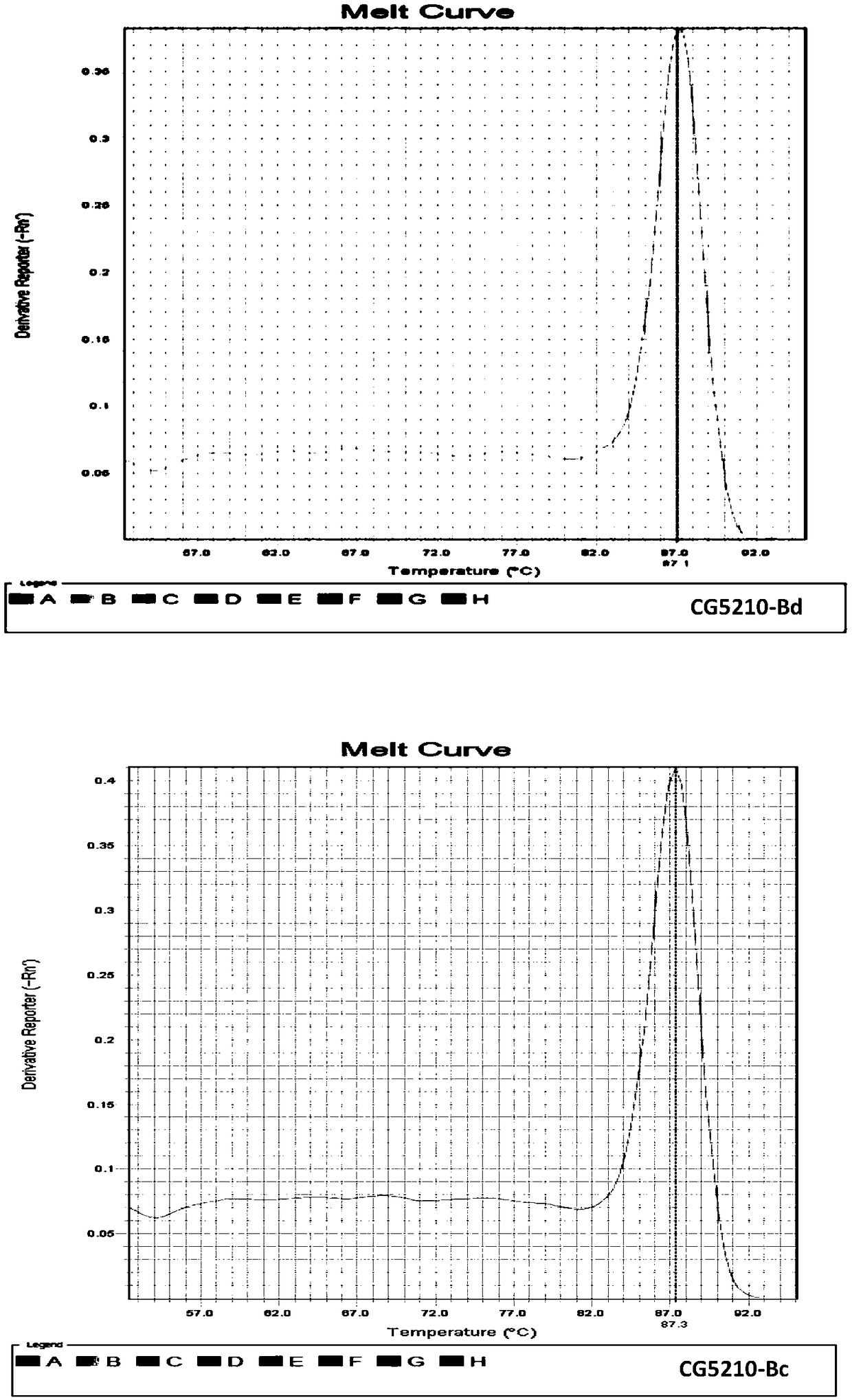
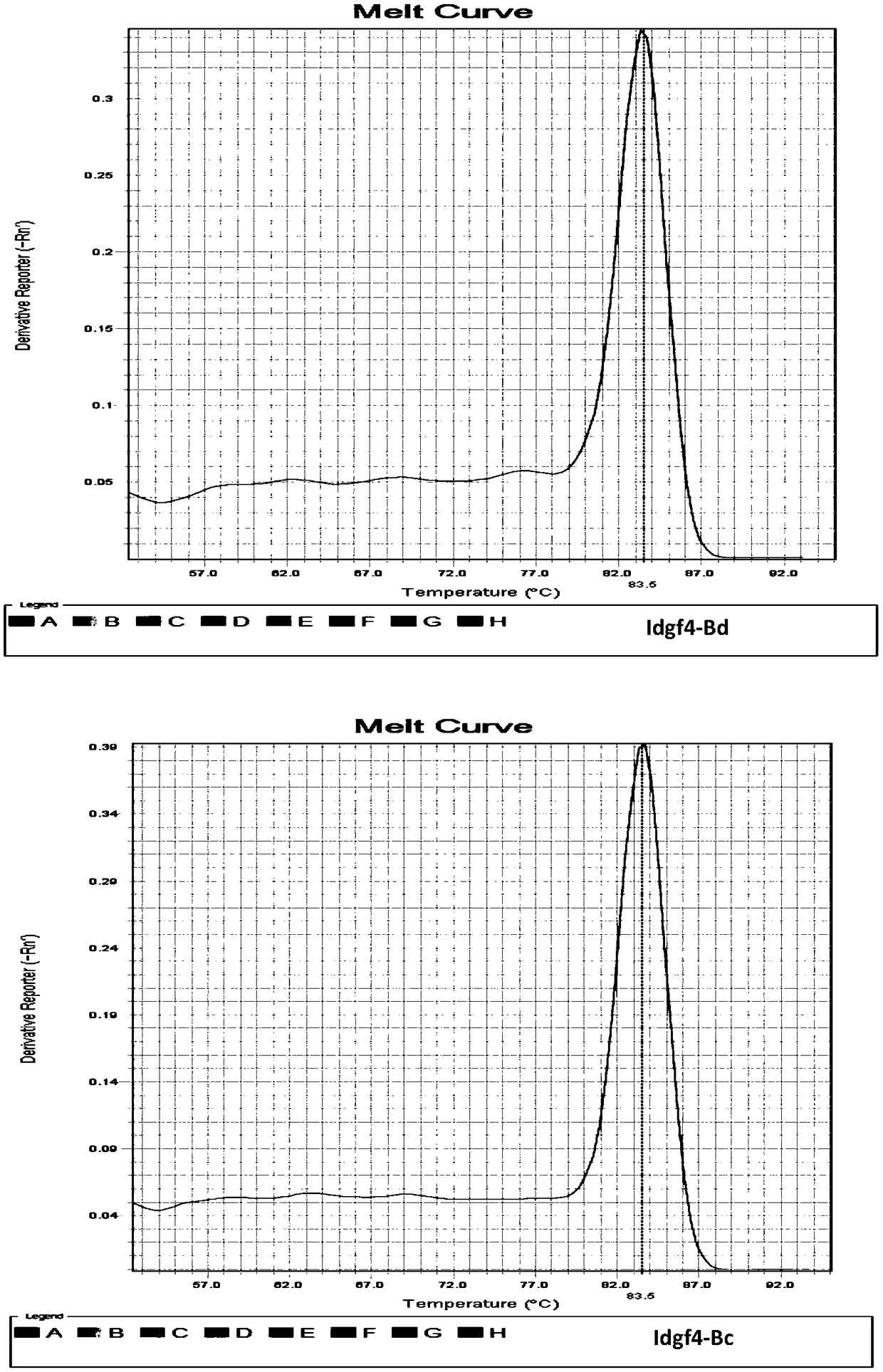
[0043] The total RNA of each fruit fly was extracted using the RNAsimple Total RNA Extraction Kit of Tiangen Biochemical Technology (Beijing) Co., Ltd., and then reverse-transcribed to obtain cDNA. Using the cDNA obtained by reverse transcription of Bactrocera dorsalis and Bactrocera guava as templates, the Takara...
Embodiment 3
[0054] The primers of embodiment 3, embodiment 1 are to the optimization of required template amount
[0055] Carry out 10-fold gradient dilution respectively to the cDNA that embodiment 2 obtains, obtain the cDNA dilution liquid of the dilution 10 times, 100 times, 1000 times and 10000 times of the specific development stage of each fruit fly (the content of cDNA is respectively 100 μ g / μl, 10 μ g / μl, μl, 1 μg / μl and 0.1 μg / μl), and then use this as a template to determine the optimal template amount using the reaction system and reaction procedure in Example 2, and use water to set up a negative control.
[0056] The results showed that using 100 μg / μl, i.e. 10-fold diluted cDNA, as a template, both primer pairs could obtain positive amplification curves in B. dorsalis and B. guava, and the amplification curves of internal reference genes and target genes The CT value of the target gene is within the reasonable range of 20-30; while 10 μg / μl, 1 μg / μl and 0.1 μg / μl cDNA as t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com