Leon-rc compression method for genome sequencing data
A genome sequencing and compression method technology, applied in the field of biological information, can solve the problems of low compression rate, long time to find anchor points, no consideration of mirror repetition, reverse repetition, etc., to achieve the effect of reducing size and size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
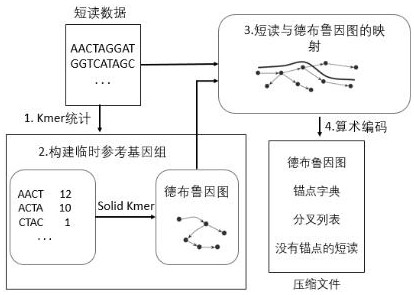
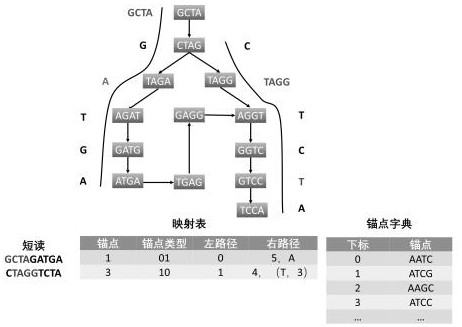
[0038] The present invention provides a LEON-RC compression method of genome sequencing data, which is mainly to improve the step of constructing an anchor dictionary for the Leon algorithm, including the following steps:
[0039] (1) divide short reading into multiple KMERs;
[0040] (2) Select a KMER to calculate its direct repetition, mirror repetition, reverse repetition, complementary repayment KMER value, compare these four values, get the smallest KMER value;
[0041] (3) Put the smallest KMER value into the Buron filter to match the lookup, and Solidkmer is stored in the Buron filter to determine if there is a minimum KMER value in the Solid Kmer; if it exists, add this to the anchor dictionary The smallest KMER value and ends the lookup; if there is no existence, get the next KMer, repeat steps (2), (3);
[0042] (4) If the smallest KMER value of all KMERs does not exist in the Solid Kmer, the short reading does not exist;
[0043] (5) Construct an anchor dictionary by st...
Embodiment 2
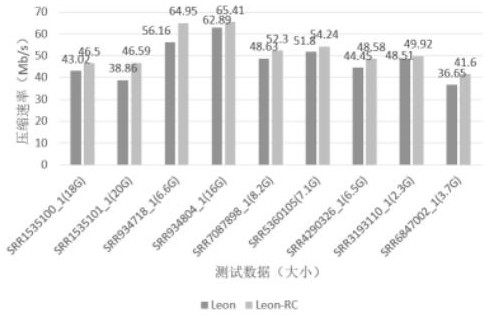
[0058] This embodiment compresses the two-generation sequencing data of different sizes, and the results of the compression test are image 3 As shown, the compression rate is significantly improved compared to Leon, Leon-RC is remarkably increasing in the case where the compression ratio is constant. Where the compression rate of the SRR934718_1 file is maximized, 56.16Mb / s is increased to 64.95MB / s. The increase is as high as 15.6%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com