Microbial data processing method for high-throughput sequencing
A technology for data processing and sequencing data, applied in the field of high-throughput sequencing quality control, it can solve the problems of not considering potentially valuable information, high false positive rate of read assignment, and achieve accurate gene expression results, reads and contigs The effect of concentrated distribution and high clustering purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
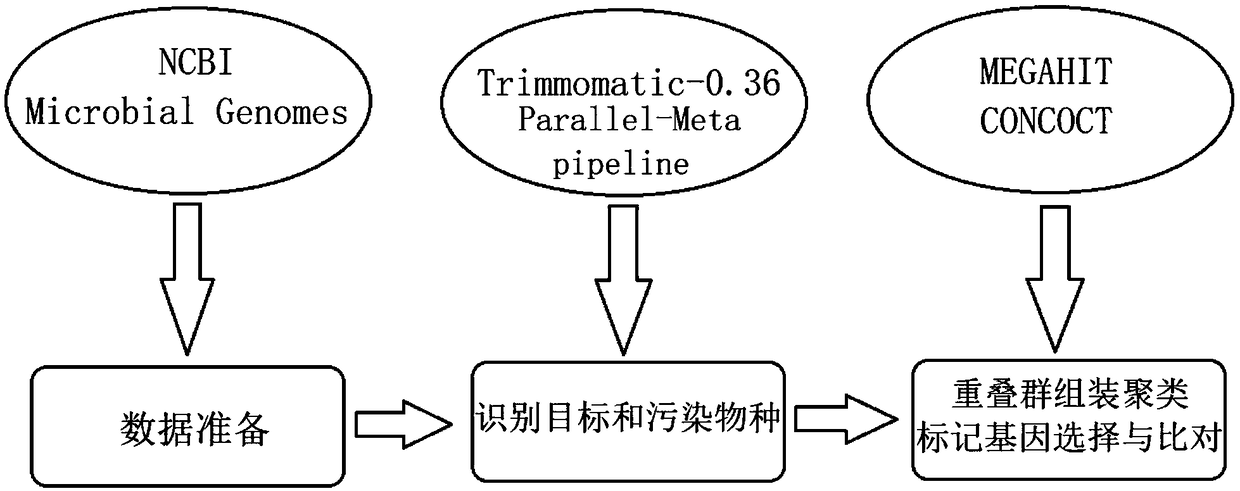
[0031] In the following, a high-throughput sequencing microbial data processing method provided by the present invention will be further described in detail and completely in conjunction with examples. The embodiments described below are exemplary, and are only used to explain the present invention, but should not be construed as limiting the present invention.
[0032] The experimental methods in the following examples are conventional methods unless otherwise specified. Unless otherwise specified, the experimental materials used in the following examples are all commercially available.
[0033] In this embodiment, the results of high-throughput sequencing of microorganisms in human sample saliva are taken as an example for quality control. The specific operation steps are as follows:
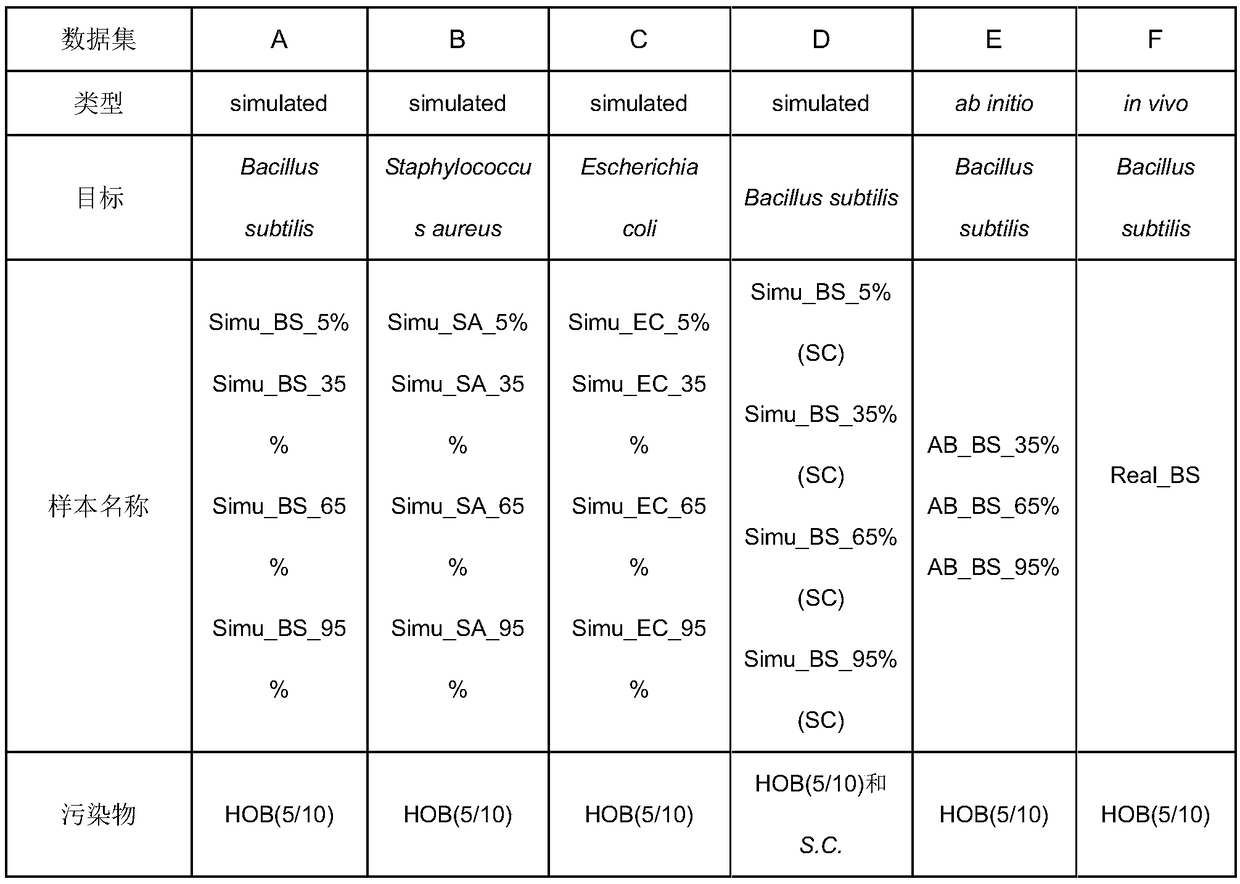
[0034] 1. Simulated and real data sets
[0035] 1. Information about simulated and real metagenomic data sets.
[0036] In this embodiment, three types of metagenomic data sets are selected: simulated...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com