Prenatal non-invasive fetal chromosome detection system based on deoxyribonucleic acid (DNA) fragment enrichment and sequencing technology
A DNA sequence and chromosome technology, applied in the field of chromosomal abnormality detection system, can solve the problems of fetal loss, inability to accurately detect chromosomal abnormalities, and low coverage depth, and achieve the effect of high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 A method for detecting prenatal fetal chromosomal abnormalities based on specific enrichment of DNA fragments, the method comprising:
[0079] (1) Obtain a peripheral blood sample from a pregnant woman, and separate the plasma free DNA in the sample;
[0080] (2) Use the DNA probes or primers of all the exons of the human genome to perform probe hybridization capture enrichment on the plasma free DNA obtained in step (1), and enrich the DNA fragments of the mother and fetus at the same time;
[0081] (3) Sequence the DNA fragments enriched in step (2), with an average sequencing depth greater than 100X, that is, more than 100 DNA fragments are covered on average on the genome sequence positions to be enriched;
[0082] (4) According to the sequencing results obtained in step (3), identify the position of each DNA sequence obtained by sequencing on the human reference genome, and for each target enrichment region on the human reference genome, the DNA sequence ...
Embodiment 2
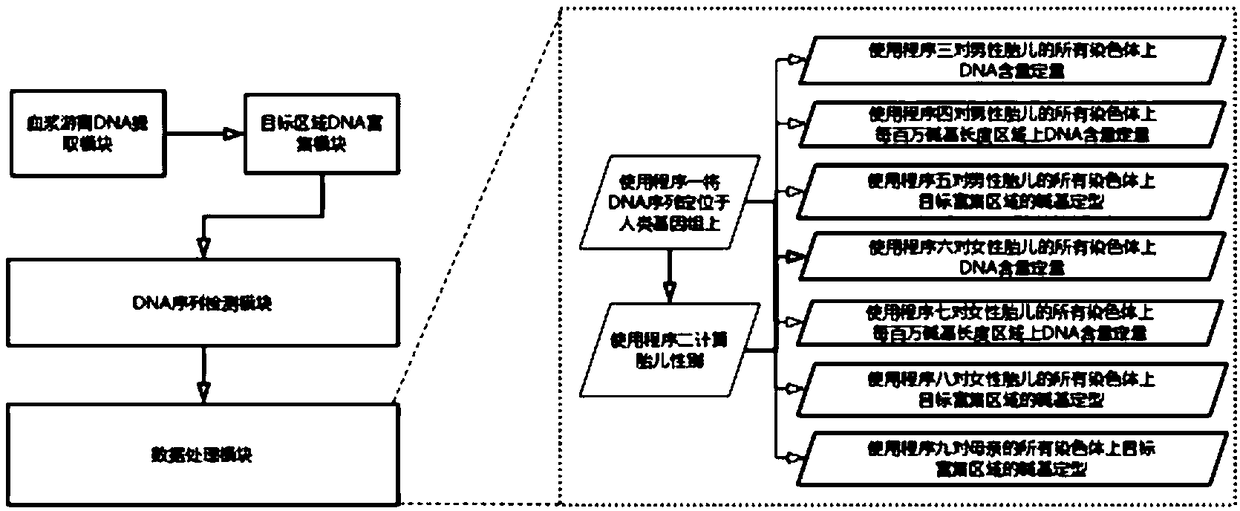
[0087] Example 2 A prenatal non-invasive fetal chromosome detection system based on DNA fragment-specific enrichment and second-generation sequencing technology, including four modules:
[0088] Module 1, obtaining peripheral blood samples from pregnant women, separating and extracting plasma free DNA in the samples, this module is a set of DNA extraction devices;
[0089] Module 2, using DNA probes or primers at specific positions on the human genome to enrich the plasma cell-free DNA obtained in module 1 to obtain DNA fragments, this module is a set of DNA enrichment devices;
[0090] Module 3: Sequence and detect the DNA fragments enriched in Module 2 to obtain the detected DNA sequence. This module is a second-generation DNA sequencer;
[0091] Module 4, data processing of the DNA sequence detected by module 3, which includes a computer and 9 calculation programs:
[0092] Program 1, this program is a sequence positioning program, which locates the sequence on the long se...
Embodiment 3
[0101] Example 3 A method for detecting prenatal fetal chromosomal abnormalities based on specific enrichment of DNA fragments, the method comprising:
[0102] (1) Obtain the peripheral blood sample of the pregnant woman, use the DNA extraction device in module 1 to separate and extract the plasma free DNA in the peripheral blood sample, and obtain the plasma free DNA;
[0103] (2) using the DNA enrichment device in module two and the DNA probe or primer on the human genome to enrich the plasma free DNA obtained in step (1) to obtain DNA fragments;
[0104] (3) Use the second-generation DNA sequencer in module three to sequence the DNA fragments obtained in step (2), to obtain a detected DNA sequence;
[0105] (4) Use computer and 9 programs to carry out data processing to the detected DNA sequence obtained in step (3):
[0106] Program 1, this program is a sequence positioning program, which locates the detected DNA sequence obtained in module 3 on the human genome, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com