Automated analysis method based on BSA gene positioning
A technology for automated analysis and gene mapping, applied in genomics, instrumentation, proteomics, etc., can solve the problems of unreliable sequencing results, single sequencing samples, and complicated processes, and achieve rich graphical display parts, simplify method processes, and improve The effect of using efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
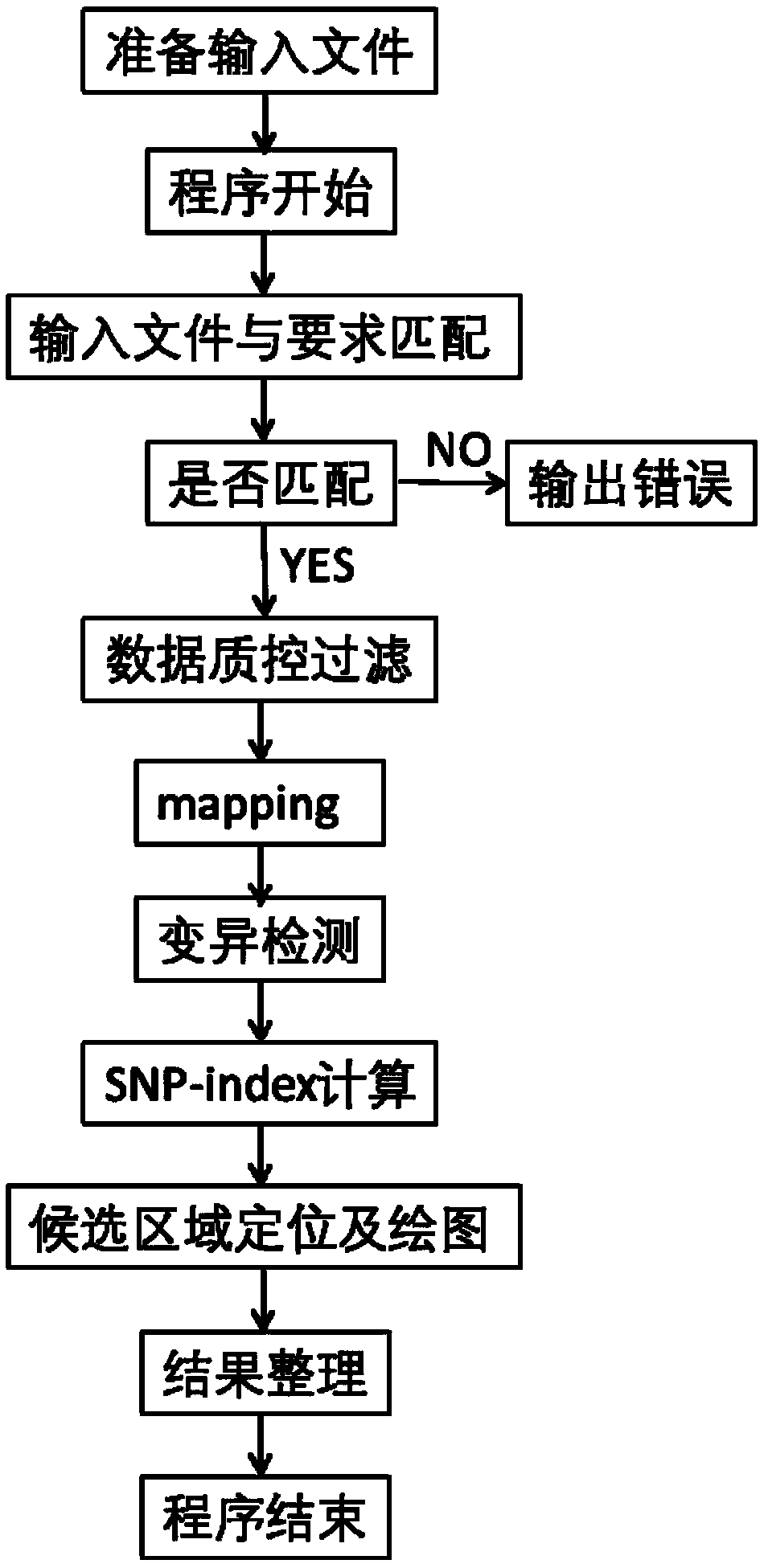
[0022] see figure 1 , a kind of automatic analysis method based on BSA gene localization shown in the figure, comprises the following steps:
[0023] (1) Prepare raw data for analysis;
[0024] (2) Detect whether the prepared file meets the conditions (file name and number of files), and judge whether it is satisfied. If it is satisfied, transfer the data to the corresponding working directory for the next step. If not, the corresponding prompt information will be output on the screen. The task corresponding to the project exits;
[0025] (3) If there is no problem in the above judgment, start to analyze the main process. If there is a problem in the process, the corresponding prompt information will be output for error correction.
[0026] In practical application, the toolkit that method of the present invention utilizes comprises 1 Shell script code altogether, and script name is as follows:
[0027] (a)BSA_Pipelines.sh
[0028] The code of this script is written based ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com