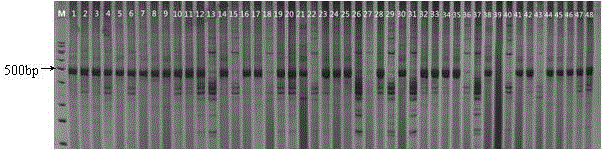
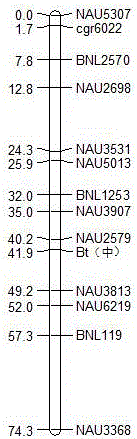
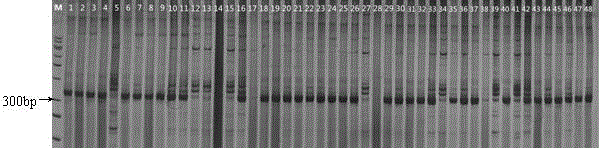
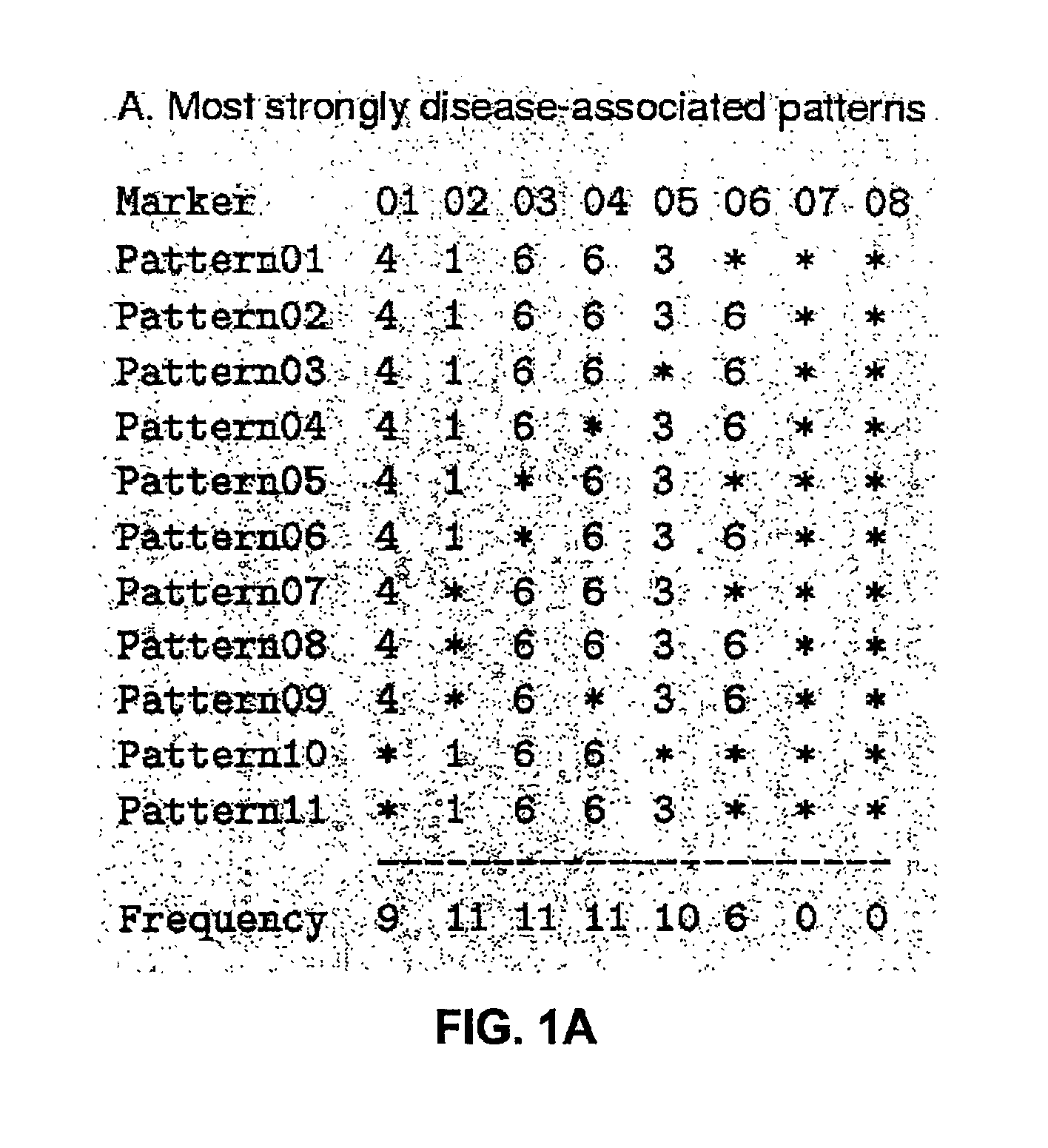
Patents
Literature
124 results about "Gene Localization" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
SNP (Single Nucleotide Polymorphism) molecular marker chained with pumpkin peel color gene and application thereof
ActiveCN106636393AShorten the timeImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationGene mappingMolecular breeding
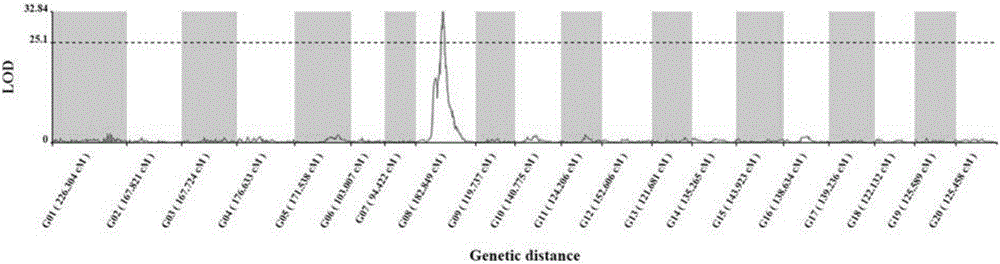
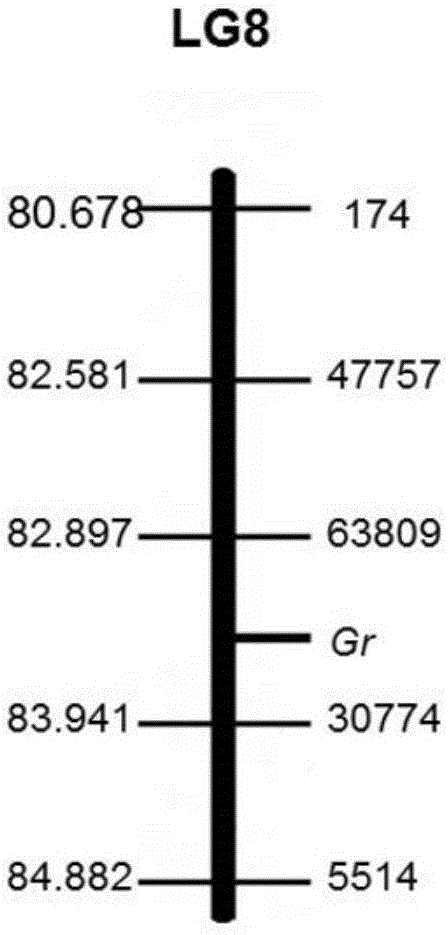
The invention discloses an SNP (Single Nucleotide Polymorphism) molecular marker chained with a pumpkin peel color gene and an application thereof. A molecular marker 63809 and a molecular marker 30774 obtained by screening are tightly chained with the pumpkin peel color gene, and the genetic distance between the two markers is 1.04cM; the SNP molecular marker can be directly applied to building of a peel color gene molecular marker assisted breeding system. A dCAPS amplification primer designed according to the two molecular markers can be used for improving molecular assisted breeding of pumpkin varieties easily, conveniently and rapidly at a high flux, technical support is provided for pumpkin appearance quality molecular breeding, and meanwhile the conventional gene mapping time is shortened greatly.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Quantitative character gene site locating method based genomic exon chip
The new quantitative character gene site locating method based on genome exon chip is to utilize exon sequence as one kind of molecular mark and utilize the detected data of human, rice and other allgenome or exon sequence chip of partial chromosome as colony locating molecular mark data for gene location via statistic location method. The present invention can obtain located functional gene sequence directly, and is suitable for all the quantitative character and complicated character of life. The research colonies include plant DH colony, F2 colony, near isogenic line, near isogenic introduction line, recombinant idiogamy line, animal family, etc.
Owner:ZHEJIANG UNIV
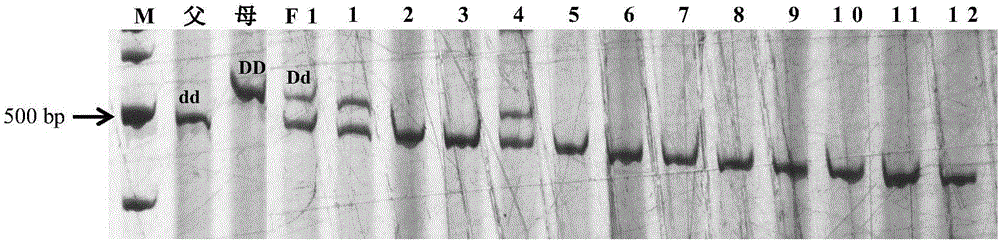
Mapping of eggplant fruit color epistatic gene D, and InDel molecular marker development and application thereof
ActiveCN106701926AImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationRe sequencingAgricultural science
The invention discloses a mapping result of an eggplant fruit color epistatic gene D. The gene D is mapped in a position near a nucleotide sequence Sme2.5_00538.1 on a No.10 chromosome of the eggplant. Based on the re-sequencing result of male and female parents, an InDel molecular marker closely linked with the eggplant fruit color epistatic gene D is obtained near the nucleotide sequence Sme2.5_00538.1 of the No.10 chromosome, and is named as InDel22522; the genetic distance of the marker from the epistatic gene D is 0.61 cM. The nucleotide sequence of the molecular marker InDel 22522 in the female parent has one 29-basic-group InDel more than that in the male parent. The mapping result on the eggplant fruit color epistatic gene D provided by the invention lays the foundation for the precise positioning and cloning of the eggplant fruit color epistatic gene; the InDel 22522 molecular marker and a molecular marker amplification primer can be applied to eggplant fruit color breeding practice in a way of simplicity, convenience, high speed and high flux; the eggplant fruit color property improving process is accelerated.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Design method of SSR label primer and wheat SSR label primers
InactiveCN103571833AIncrease the number ofFast encryptionMicrobiological testing/measurementSpecial data processing applicationsContigTest group
The invention discloses a design method of an SSR label primer. In order to overcome the defect of insufficient polymorphism of the conventional SSR label primer, the invention provides a method for designing a novel SSR label primer based on a draft sequence of a genome. The design method comprises the following steps: firstly selecting an SSR label primer with a known site as a starting SSR label primer; secondly, comparing the starting SSR label primer with the draft sequence of a chromosome to which the starting SSR label primer belongs, and finding a contig in a comparison result; thirdly, searching an SSR sequence in the contig as a finishing SSR sequence; finally, designing the SSR label primer based on the finishing SSR sequence. The invention provides 14 pairs of novel SSR label primers related to wheat stripe rust resistance, and a method for establishing a wheat genetic map with L693*L661 and L661*L693F2 single plants as a mapping population, wherein five pairs of the SSR label primers generate genetic polymorphism in a genetic test group. According to the design method, the number of the SSR label primers with known sites can be quickly increased and the polymorphism of the SSR label is increased; the genetic map can be quickly encrypted through combination with initial gene localization.
Owner:SICHUAN AGRI UNIV
Rapid positioning method of cotton single locus quality gene in chromosome
ActiveCN105925668AReduce workloadImprove positioning efficiencyMicrobiological testing/measurementPopulationCoincidence
The invention relates to a rapid positioning method of cotton single locus quality gene in chromosome, and belongs to the technical field of biology. The method includes establishing a near-isogenic mixed pool by using land-sea F2 positioning group, screening the gene to obtain a primer with polymorphism, detecting the DNA of 9 single strains, P-1, P-2 and F1 in a recessive gene mixed pool, recording the banding patterns of the DNA, calculating the individual number among the 9 single strains in the recessive mixed pool which has the same banding pattern with the recessive parent P-1, calculating the coincidence rate, selecting the polymorphic primer having the banding pattern coincidence rate over 60% with P-1 and having dominant or codominant F1 banding pattern, detecting the F2 segregation population by using the small amount of the primer, calculating the banding pattern, and analyzing by software to obtain the SSR primer concatenated with the quality gene. Displayed by the embodiment, the method makes the range of screening target concatenated primer by using F2 positioning group reduce over 90%, can lock the SSR primer concatenated with the target quality gene rapidly, can reduce the workload of the gene positioning effectively, and can improve the positioning efficiency of the quality gene in the chromosome greatly.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
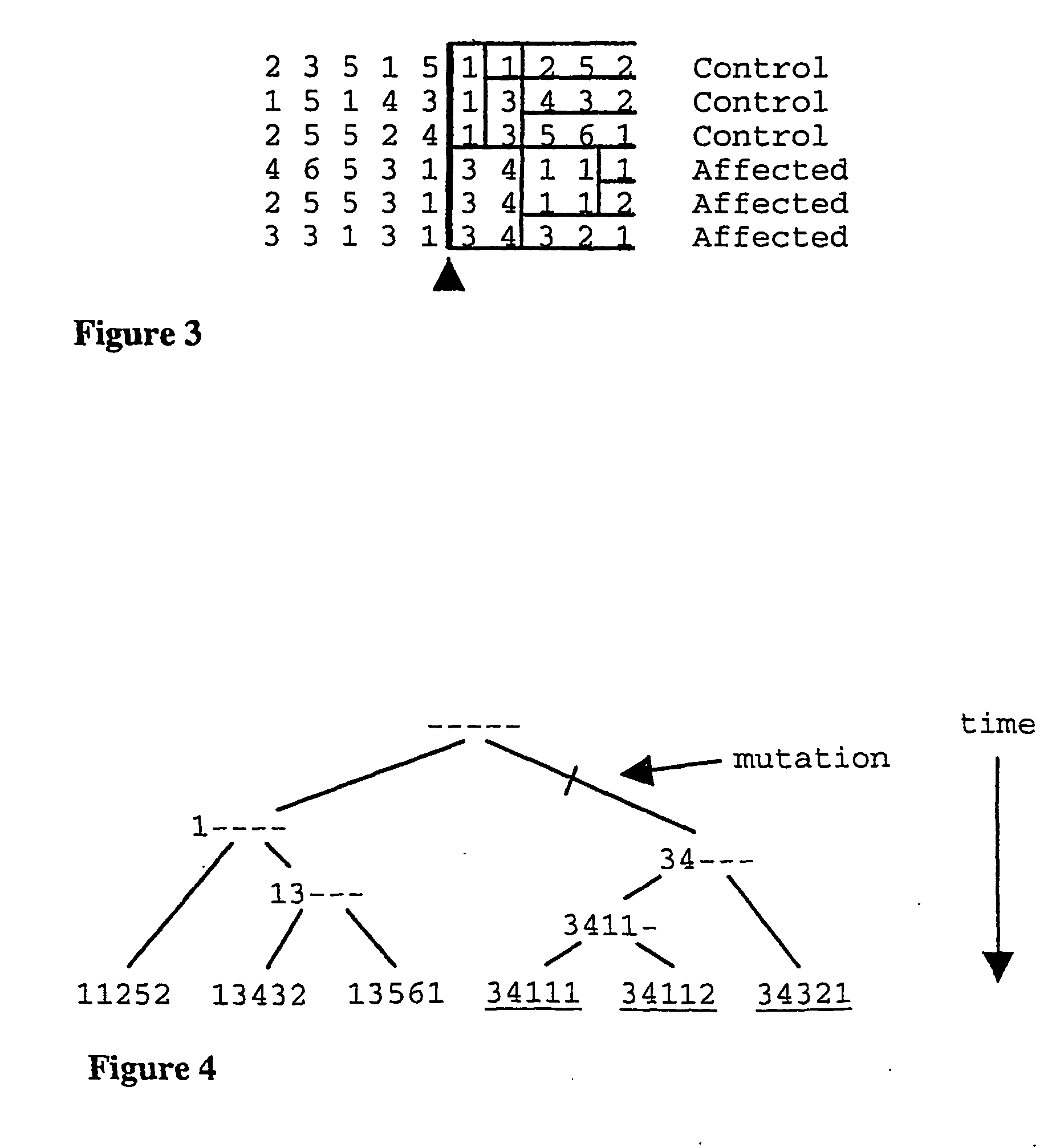
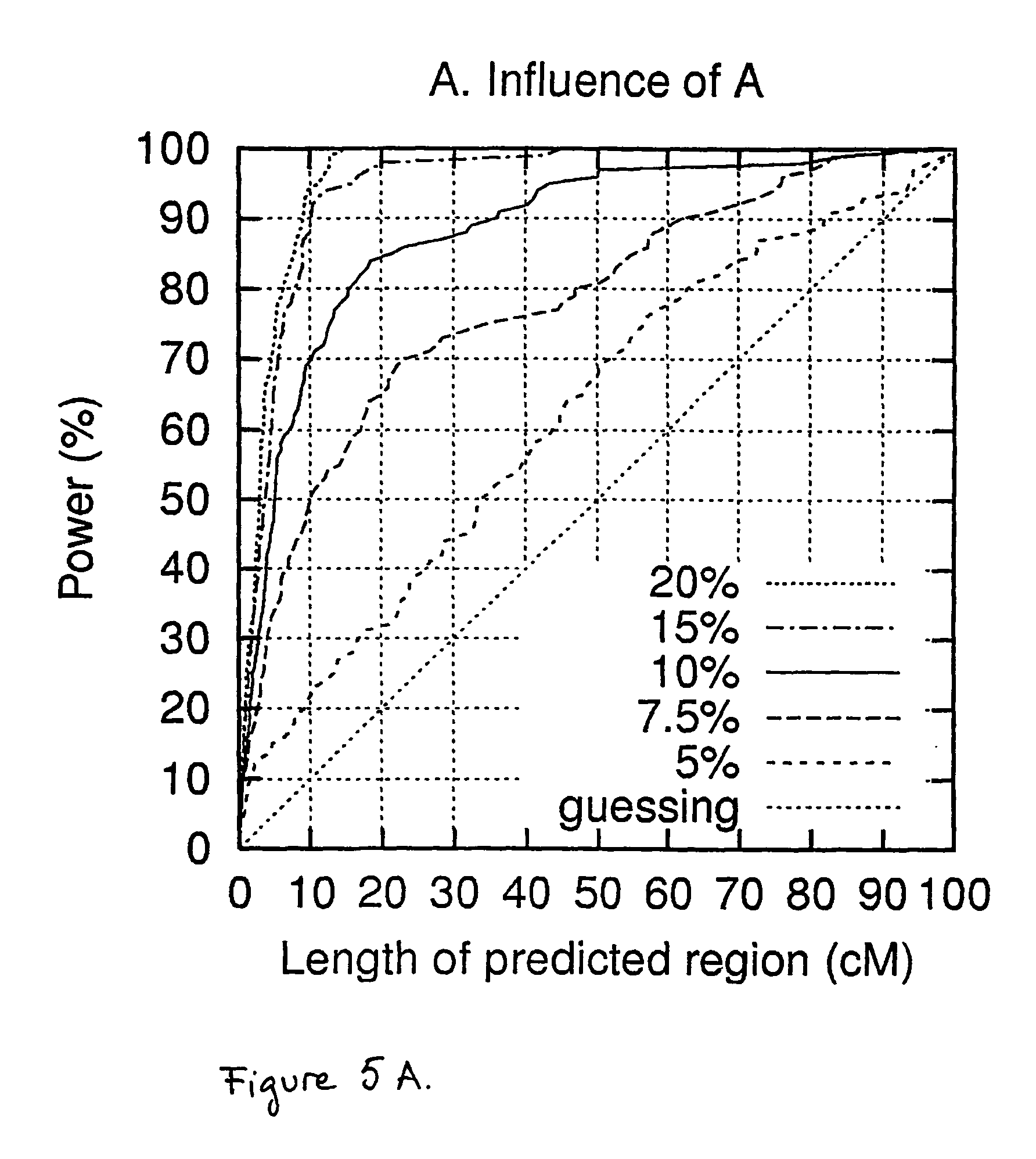
Method for gene mapping from chromosome and phenotype data
InactiveUS6909971B2Data processing applicationsMicrobiological testing/measurementGene mappingNucleotide
The present invention relates to a method for gene mapping from chromosome and phenotype data, which utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
Fluorescent labeled X-STR gene locus multiplex PCR method and application thereof
InactiveCN102250883AHigh polymorphismHigh application valueMicrobiological testing/measurementDNA preparationMolecular biologyGene Localization
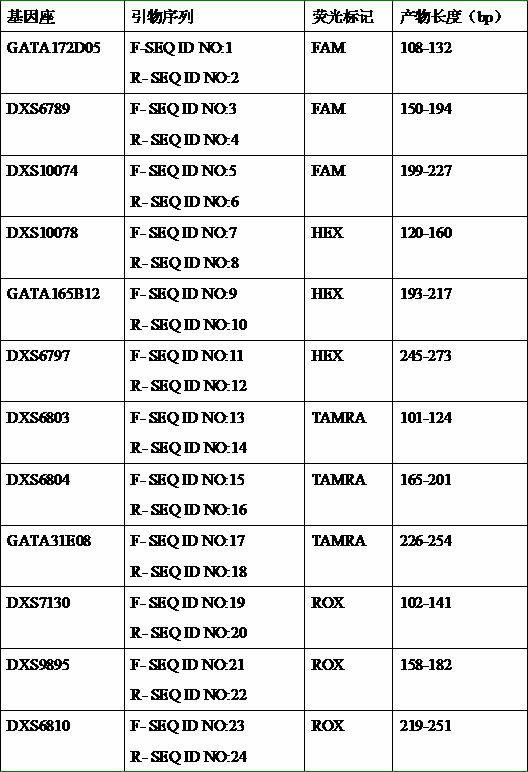
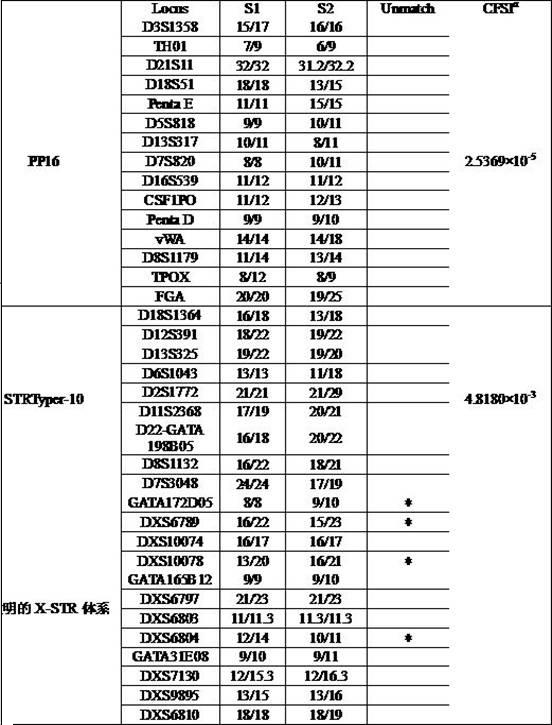
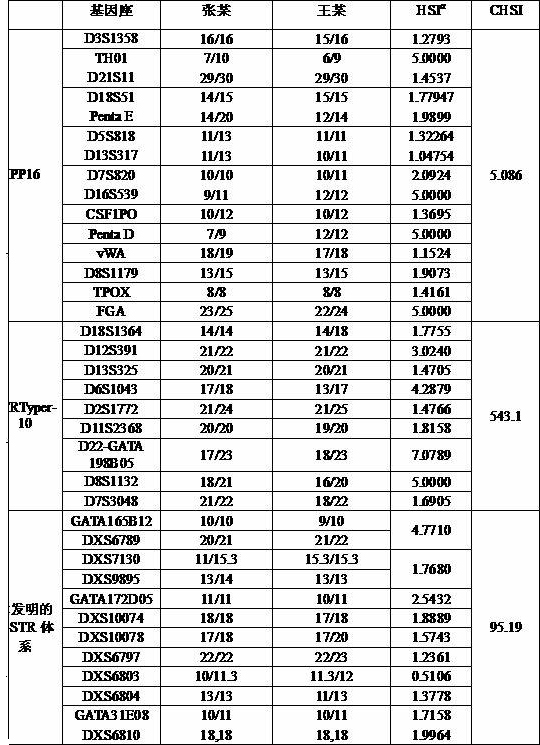
The invention discloses a fluorescent labeled X-STR locus multiplex PCR method and an application thereof; the system performs multiplex amplification analysis of 12 loci: GATA172D05, DXS6789, DXS10074, DXS10078, GATA165B12, DXS6797, DXS6803, DXS6804, GATA31E08, DXS7130, DXS9895, and DXS6810, wherein primers of the 12 loci are labeled respectively by four fluorescences of FAM, HEX, TAMRA, ROX. The method of the invention can be used to prepare a set of kit as an X-STR locus multiplex amplification kit with Chinese characteristics, and the kit is applicable to gene localization of paternity identification, individual identification, sex identification, and X-linked genetic diseases, and is especially applicable to paternity identification for sister recognition, half-blood half-sister recognition, generation-skipping recognition, etc.
Owner:SUN YAT SEN UNIV
Lancea tibetica microsatellite molecular markers
The invention relates to Lancea tibetica microsatellite molecular markers, belonging to the technical field of DNA (deoxyribonucleic acid) molecular markers. The Lancea tibetica microsatellite molecular markers are prepared by the following steps: separating the microsatellite molecular markers from the Lancea tibetica genome DNA by a RAD-seq technique, designing specific primers of each molecular marker are designed according to the flanking sequences on both ends of the site of the microsatellite molecular marker, carrying out PCR (polymerase chain reaction) amplification on the Lancea tibetica individuals from different populations, and detecting the stability and polymorphism of the amplification resulti, thereby obtaining 10 effective microsatellite molecular markers. The invention develops the effective Lancea tibetica microsatellite molecular markers, and establishes the method for preparing the Lancea tibetica microsatellite molecular markers. The Lancea tibetica microsatellite molecular markers can be used for genetic diversity analysis, plant genetic map establishment, gene localization, variety identification, germplasm preservation, quantitative character gene analysis, evolution, genetic relationship research and the like.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
Method for gene mapping from genotype and phenotype data
InactiveUS20050250098A1Model-free and computationally effectiveOvercomes drawbackMicrobiological testing/measurementBiostatisticsGene mappingGenetic linkage disequilibrium
A method for gene mapping from genotype and phenotype data utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. All marker patterns P that satisfy a certain pattern evaluation function e(P) are searched from the data, each marker mi of the data is scored by a marker score and the location of the gene is predicted as a function of the scores s(mi) of all the markers mi in the data.
Owner:LICENTIA OY
Molecular marker method of cucumber spire etiolation gene
ActiveCN106957897AAccurate genetic lociEasy to identifyMicrobiological testing/measurementBiotechnologyGene mapping
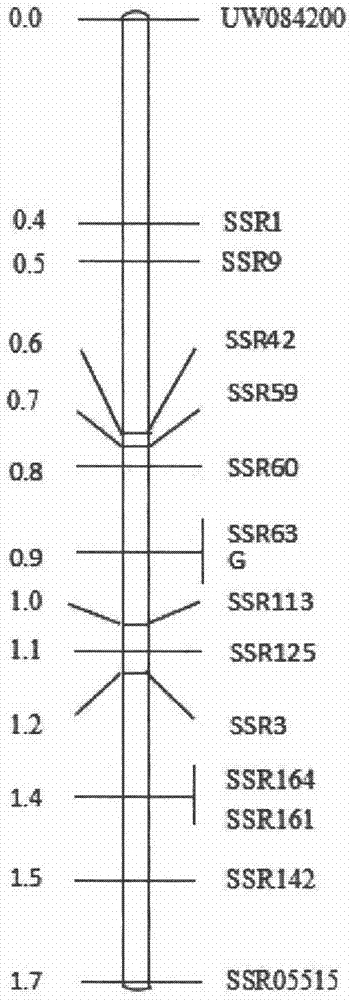
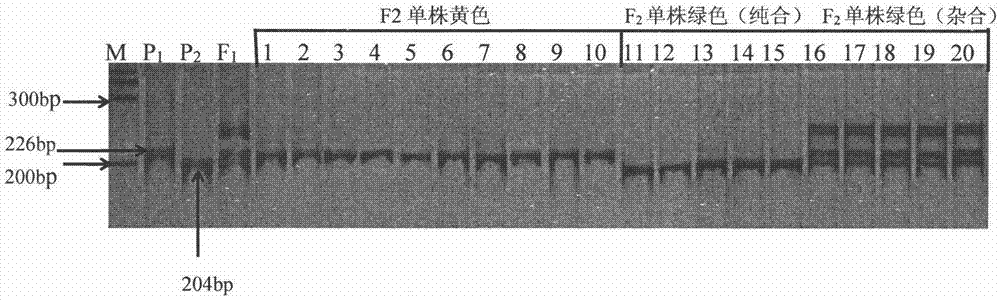
The invention discloses a molecular marker method of cucumber spire etiolation gene linking, belonging to the biotechnical field. A molecular marker SSR63 closely linked to the cucumber spire etiolation gene is disclosed for the first time, the forward and reverse primers of the marker are respectively: 5'GCTTCATCCTGTGGCGT3' and 5'CCAGTCATGGGTTTGTTGGA3'. In F2 constructed from spire cucumbers and green-leaf cucumbers, 226bp products can be amplified from the primers SSR63F / SSR63R inspire etiolation strain, 204bp product or product containing both 204bp and 226bp strips can be amplified in green-leaf strain, and all the products can be closely linked with vyl. The molecular marker closely linked with cucumber spire etiolation gene vyl can be used for cucumber spire etiolation gene mapping or molecular marker-assisted breeding.
Owner:NANJING AGRICULTURAL UNIVERSITY
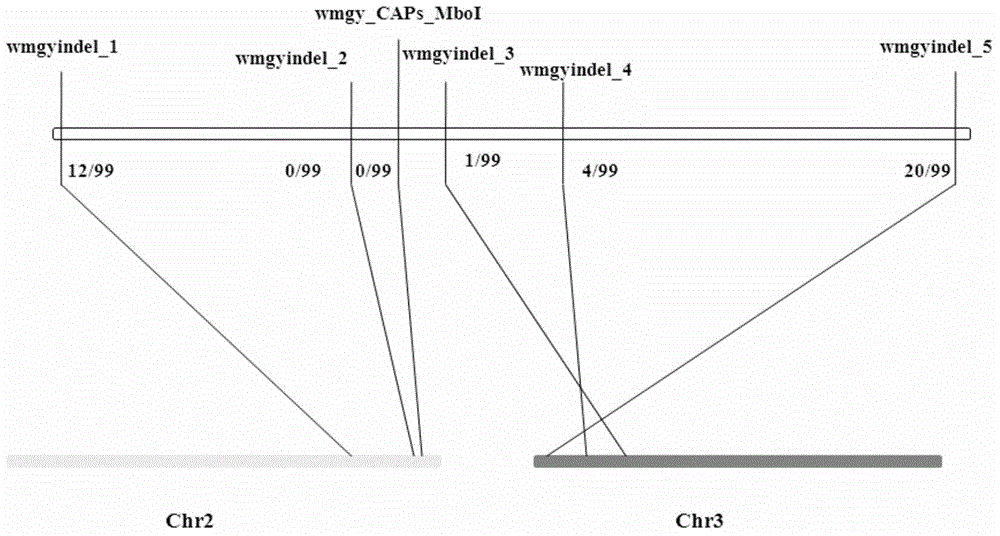
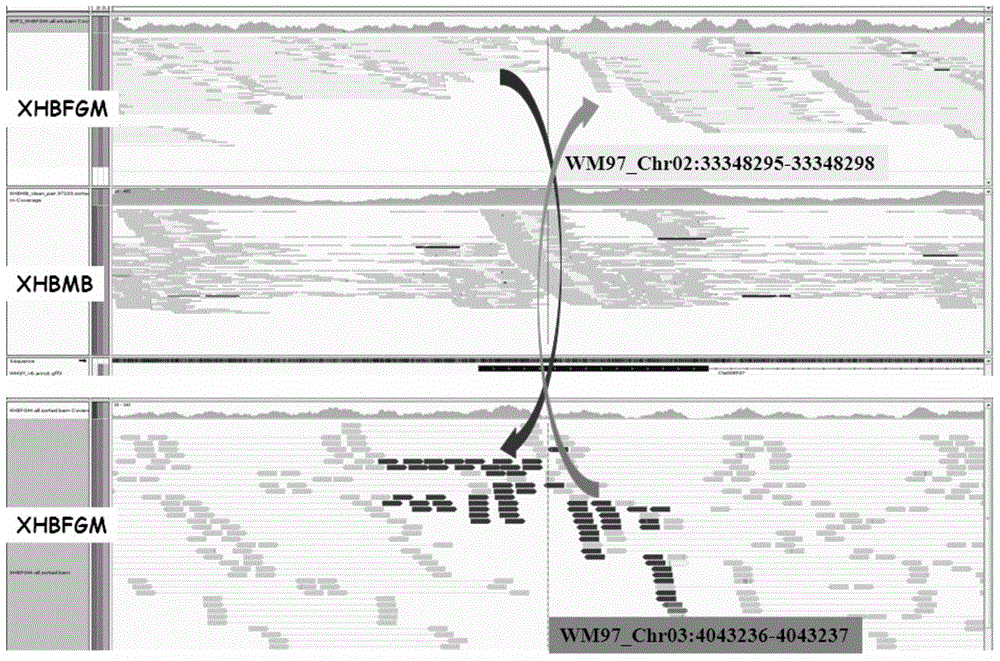
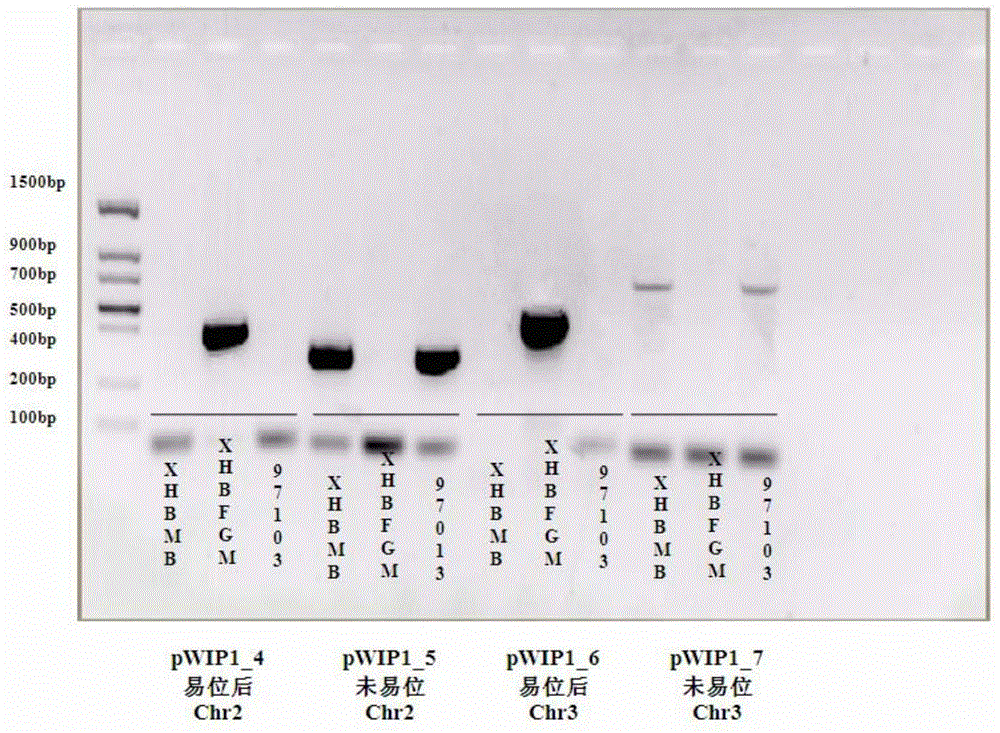
Watermelon female lines gene C1WIP1 and chromosome translocation and linkage marker
ActiveCN103937785AFast and accurate cloningReduce the cost of seed productionMicrobiological testing/measurementFermentationGenome resequencingA-DNA
The invention belongs to the molecular biology field, and more specifically relates to watermelon female lines gene(i) C1WIP1( / i) and chromosome translocation and linkage marker, which comprises a DNA sequence of watermelon female lines gene(i) gy( / i) gene(i) C1WIP1( / i), its acquisition method, a molecular marker linked with the DNA sequence, a chromosome translocation section due to appearance of the watermelon female lines and its acquisition method. According to the invention, a classic gene positioning method and a whole genome re-sequencing technology are organically combined, and watermelon female lines gene(i) C1WIP1( / i) is rapidly and accurately cloned; a PCR technology is used for confirming that a watermelon spontaneous mutant material XHBFGM is appeared by female lines phenotype due to chromosome translocation, and a preliminary research is carried out on the effect mechanism of (i) C1WIP1( / i); By aiming at the existence of the chromosome translocation, the molecular marker linked with watermelon female lines gene(i) C1WIP1( / i) is designed, the watermelon female lines phenotype which takes XHBFGB as a transfer material can be rapidly detected, the molecular marker is used for assistant breeding, seed production cost of watermelon is greatly reduced, and the seed purity is increased.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
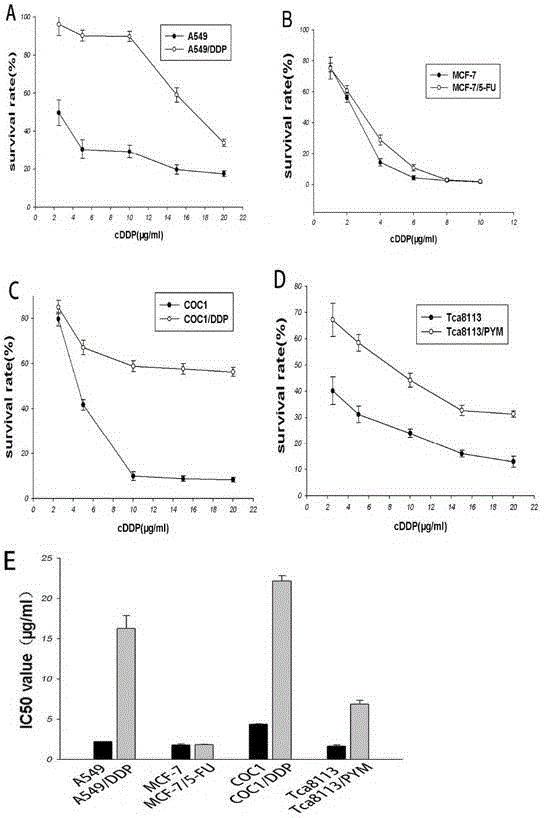
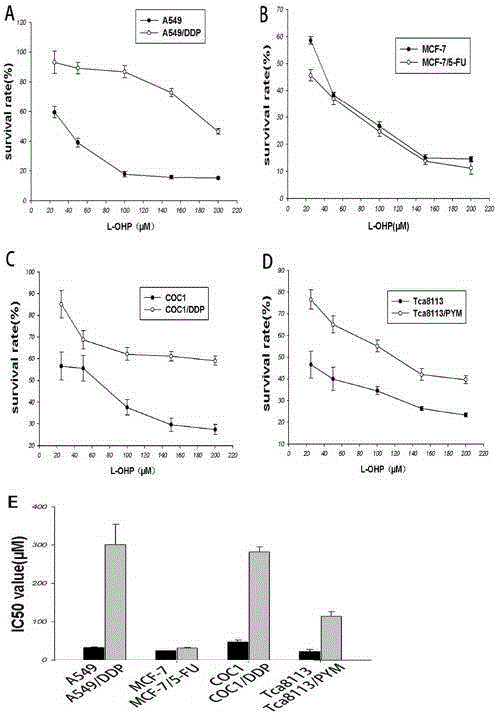
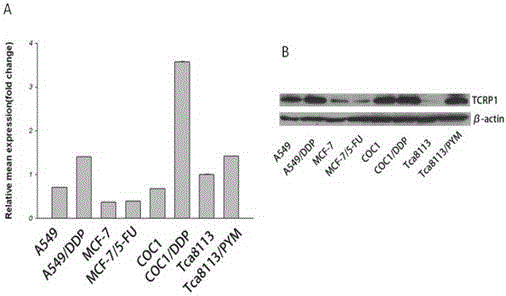
Application of TCRP1 gene in the preparation of tumor cell platinum resistance reversal agent
ActiveCN103055326AIncreased sensitivityGood curative effectPeptide/protein ingredientsGenetic material ingredientsPlatinum resistanceOpen reading frame
The invention discloses a self-cloning new human tumor drug-resistant related gene, namely TCRP1, wherein the gene and drug resistance generation of tumor chemotherapy platinum drugs has confidential relation. The expression of TCRP1 in platinum secondary drug-resistant tumor cell strains such as Tca8113 / PYM, A549 / DDP, Coc1 / DDP is raised, and the expression of TCRP1 in primary platinum drug-resistant cell strain such as lung cancer 95D is obviously superior to that of other sensitive cell strains of lung cancer. The gene is located in human chromosome 11q13.4, wherein the total length of sequence is 1834bp, the open reading frame is 708bp, and 235 amino acids are encoded. The application not only provides new information for tumor drug-resistant mechanism, but also provides new target for the design of anti-tumor drug-resistant drugs.
Owner:CANCER CENT OF GUANGZHOU MEDICAL UNIV
Indel molecular marker closely linked with pumpkin photoperiod insensitivity and application of indel molecular marker
ActiveCN108048595AShorten the timeHigh contribution rateMicrobiological testing/measurementAngiosperms/flowering plantsAgricultural scienceMolecular breeding
The invention discloses an indel molecular marker closely linked with pumpkin photoperiod insensitivity and application of the indel molecular marker, and belongs to the technical field of molecule detection. The indel molecular marker SEQ7593 is located on a tenth chromosome of a Chinese pumpkin, and is 280bp in size. The nucleotide sequence of the indel molecular marker is shown as SEQ ID NO.1.The indel molecular marker SEQ7593 can be directly used for creating an assistant breeding system of photoperiod insensitivity character molecular markers. Primer amplification designed according to the indel molecular marker can be applied to assistant breeding of pumpkin breed improvement molecules, technical support is provided for pumpkin photoperiod insensitivity molecular breeding, and timefor conventional gene positioning is shortened greatly.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Molecular marker, primers and method for estimating left-end length of N introgressed segment of tobacco
ActiveCN105200052AReduce the burden onReduce outputMicrobiological testing/measurementDNA/RNA fragmentationGene mappingNicotiana tabacum
The invention relates to a molecular marker, primers and a method for estimating the left-end length of an N introgressed segment of tobacco. The molecular marker is a sequence represented as Seq ID No.1 or Seq ID No2. According to estimating method, PCR (polymerase chain reaction) amplification is performed by adopting a GL4.06 primer pair and a GL3.50 primer pair as the primers and adopting to-be-detected N introgressed segment containing tobacco genome DNA (deoxyribonucleic acid) as a template, then electrophoresis detection is performed, and if a DNA segment with the corresponding size is obtained through amplification, the N introgressed segment of detected tobacco is as long as a disease-resistant control material Samsun NN in the molecular marker position; if the DNA segment with the corresponding size is not obtained through amplification, the N introgressed segment of the detected tobacco is shorter than the disease-resistant control material Samsun NN in the molecular marker position. The method can be simply, conveniently and rapidly applied to anti-TMV (tobacco mosaic virus) gene mapping of the tobacco and TMV-resistant tobacco variety breeding in a high-throughput manner, and reduction of linkage redundancy with the N gene is facilitated.
Owner:YUNNAN ACAD OF TOBACCO AGRI SCI
Molecular marker, primers and method for estimating right-end length of N introgressed segment of tobacco
ActiveCN105200149AReduce the burden onReduce outputMicrobiological testing/measurementDNA/RNA fragmentationNicotiana tabacumTobacco mosaic virus
The invention relates to a molecular marker, primers and a method for estimating the right-end length of an N introgressed segment of tobacco. The molecular marker is a sequence represented as Seq ID No.1 or Seq ID No2. According to estimating method, PCR (polymerase chain reaction) amplification is performed by adopting a TN5.51 primer pair and a TN5.34 primer pair as the primers and adopting to-be-detected N introgressed segment containing tobacco genome DNA (deoxyribonucleic acid) as a template, then electrophoresis detection is performed, and if a DNA segment with the corresponding size is obtained through amplification, the N introgressed segment of detected tobacco is as long as a disease-resistant control material Coker176 in the molecular marker position; if the DNA segment with the corresponding size is not obtained through amplification, the N introgressed segment of the detected tobacco is shorter than the disease-resistant control material Coker176 in the molecular marker position. The method can be simply, conveniently and rapidly applied to anti-TMV (tobacco mosaic virus) gene mapping of the tobacco and anti-TMV tobacco variety breeding in a high-throughput manner, and reduction of linkage redundancy with the N gene is facilitated.
Owner:YUNNAN ACAD OF TOBACCO AGRI SCI
Photoperiod-insensitivity Hd1 allele and molecular marker and application thereof
ActiveCN109055395AIncrease photosensitivityImprove adaptabilityMicrobiological testing/measurementPlant peptidesGenotypeAllele
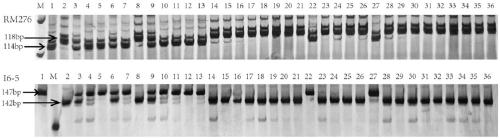
The invention discloses a photoperiod-insensitivity Hd1 allele and a molecular marker and application thereof. Zhengeng 2400 (female) of a photoperiod-insensitivity variety and Jiahe 218 (male) of a photoperiod-sensitivity variety are hybridized to obtain the genotype of an F2 single plant, and genetic linkage analysis is carried out to judge whether photoperiod-insensitivity phenotype separationof a corresponding F2:3 family is achieved or not; a heading period photoperiod-insensitivity gene of the Zhengeng 2400 is positioned at the sixth chromosome and located between a mark RM276 and a mark I6-5; the Zhengeng 2400 and the Jiahe 218 are subjected to WGS-variation point detection and gene sequencing analysis, and the analysis discovers that 123bp is inserted at the first exon 517bp of the Hd1 allele of the Zhengeng 2400; and the molecular marker H1 is developed according to the difference for detecting whether the Zhengeng 2400 and derivative varieties thereof contain the photoperiod-insensitivity gene or not. Thus, the photosensitivity of japonica rice varieties can be improved, and the regional adaptability and breeding efficiency of the japonica rice varieties are greatly improved.
Owner:ZHENJIANG AGRI SCI INST JIANGSU HILLY AREAS
Physalis angulata cpSSR-labeled primer developed based on chloroplast genome sequence and its application
ActiveCN110195123AAdd raw dataEvaluate polymorphismMicrobiological testing/measurementDNA/RNA fragmentationGene mappingGenetic diversity
The invention discloses a Physalis angulata cpSSR-labeled primer developed based on a chloroplast genome sequence and its application. The primer pair SEQ ID NO. 1 to SEQ ID NO. 60 is based on the development of the Physalis angulata chloroplast genome sequence, based on the sequencing of the chloroplast genome, a large amount of sequence information is analyzed, and then cpSSR site screening andcpSSR-labeled primer design are carried out. The patent overcomes the problem of low efficiency, long cycle and high cost of traditional SSR marker development, and fills the blank of the current cpSSR labeling information of Physalis angulata. The polymorphism and applicability of the cpSSR-labeled primer are evaluated by screening cpSSR primers and genetic diversity analysis. The cpSSR polymorphism-labeled primer provided in the patent lays the foundation for the study of genetic diversity evaluation, germplasm identification, importance gene mapping and molecular marker-assisted breeding ofPhysalis angulata.
Owner:HANGZHOU NORMAL UNIVERSITY
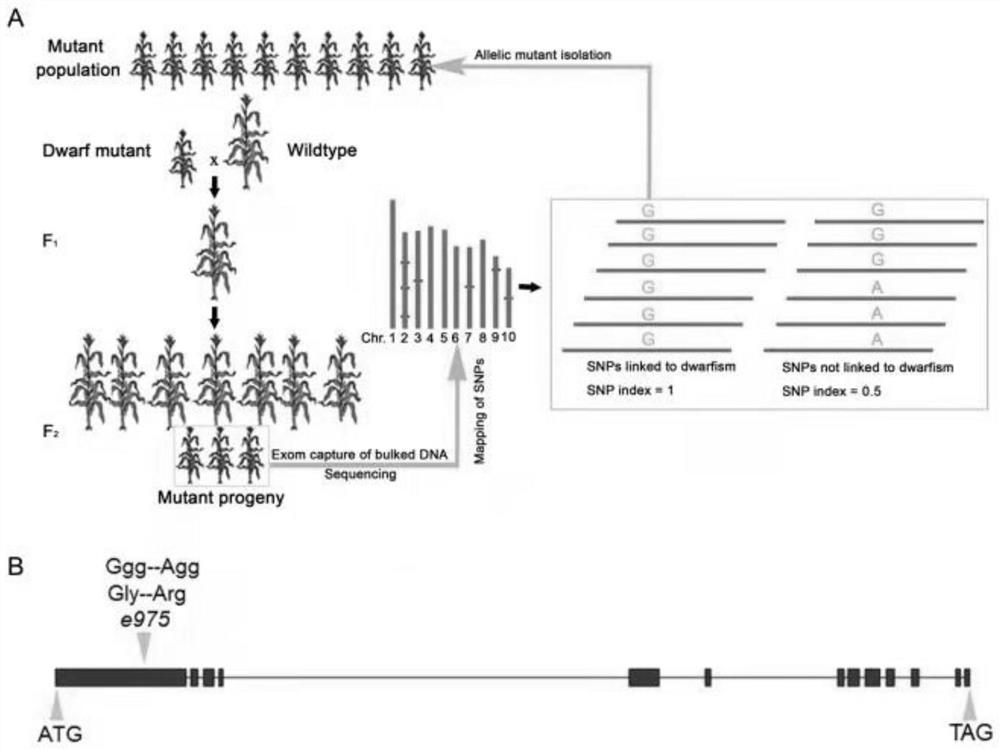
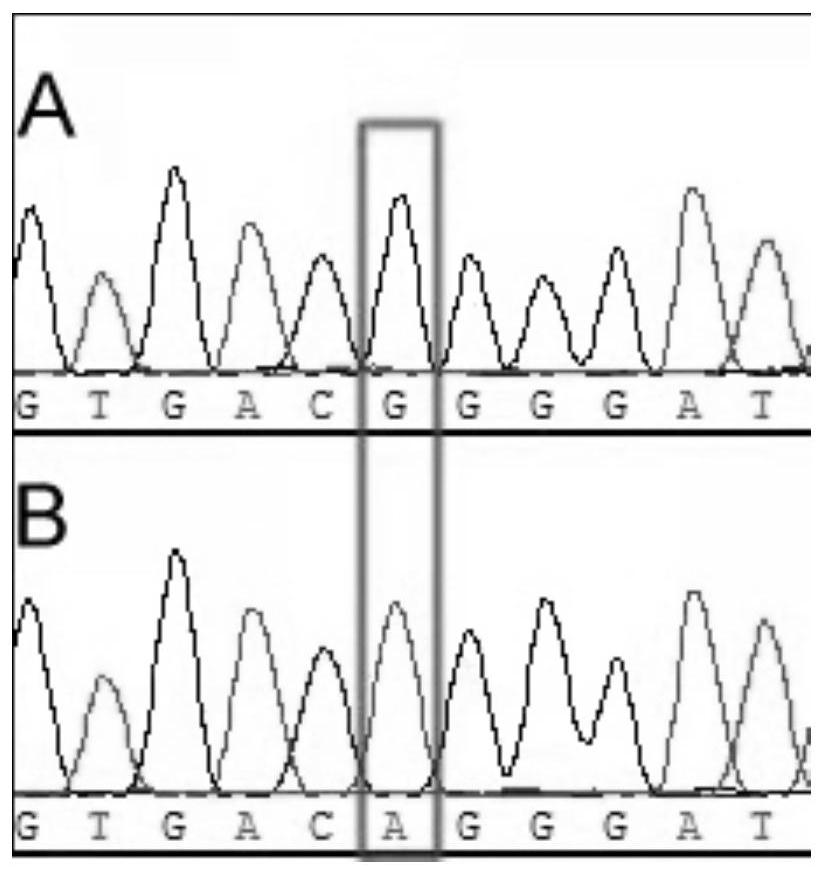
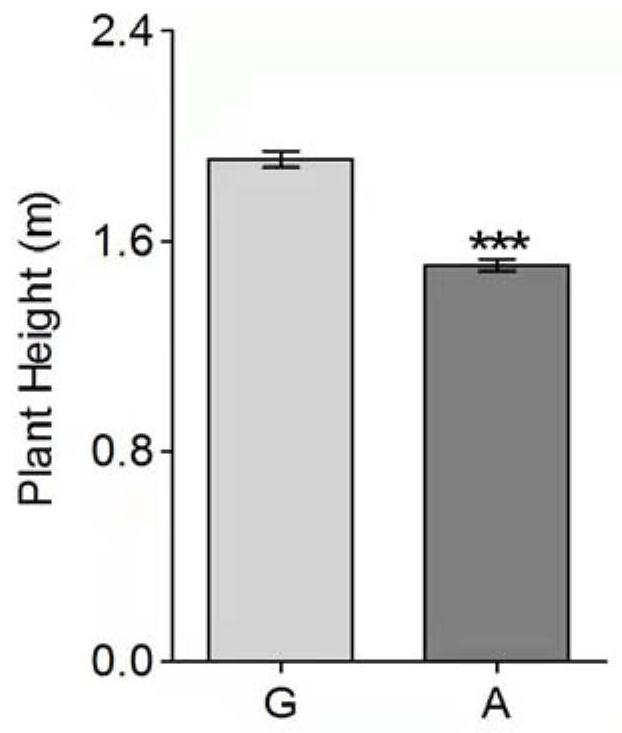
Molecular marker related to dwarfing of corn plant
ActiveCN113481315ARapid identificationShorten the breeding cycleMicrobiological testing/measurementClimate change adaptationBiotechnologyArginine
The invention discloses a molecular marker related to dwarfing of corn plant. According to the invention, a mutant e975 related to dwarfing of corn plant height is screened, although the section number of corn is obviously reduced, the height of the corn is obviously reduced, and the ear length of the corn is obviously reduced, the yield of the e975 is not obviously different from that of a wild type. Gene localization shows that the first exon of the Zm00001d027807 gene of the mutant has changed from G to A, so that non-polar glycine is changed into alkaline arginine. For the variation, a corresponding molecular marker primer (SEQ ID NO.3-4) is developed, and after PCR amplification, Sanger sequencing is utilized for genotype interpretation, so that the wild genotype and the mutant genotype of the filial generation contain corn dwarfing gene Zm00001d027807 can be identified. The molecular marker is beneficial to auxiliary breeding of new corn dwarfing varieties.
Owner:QILU NORMAL UNIV
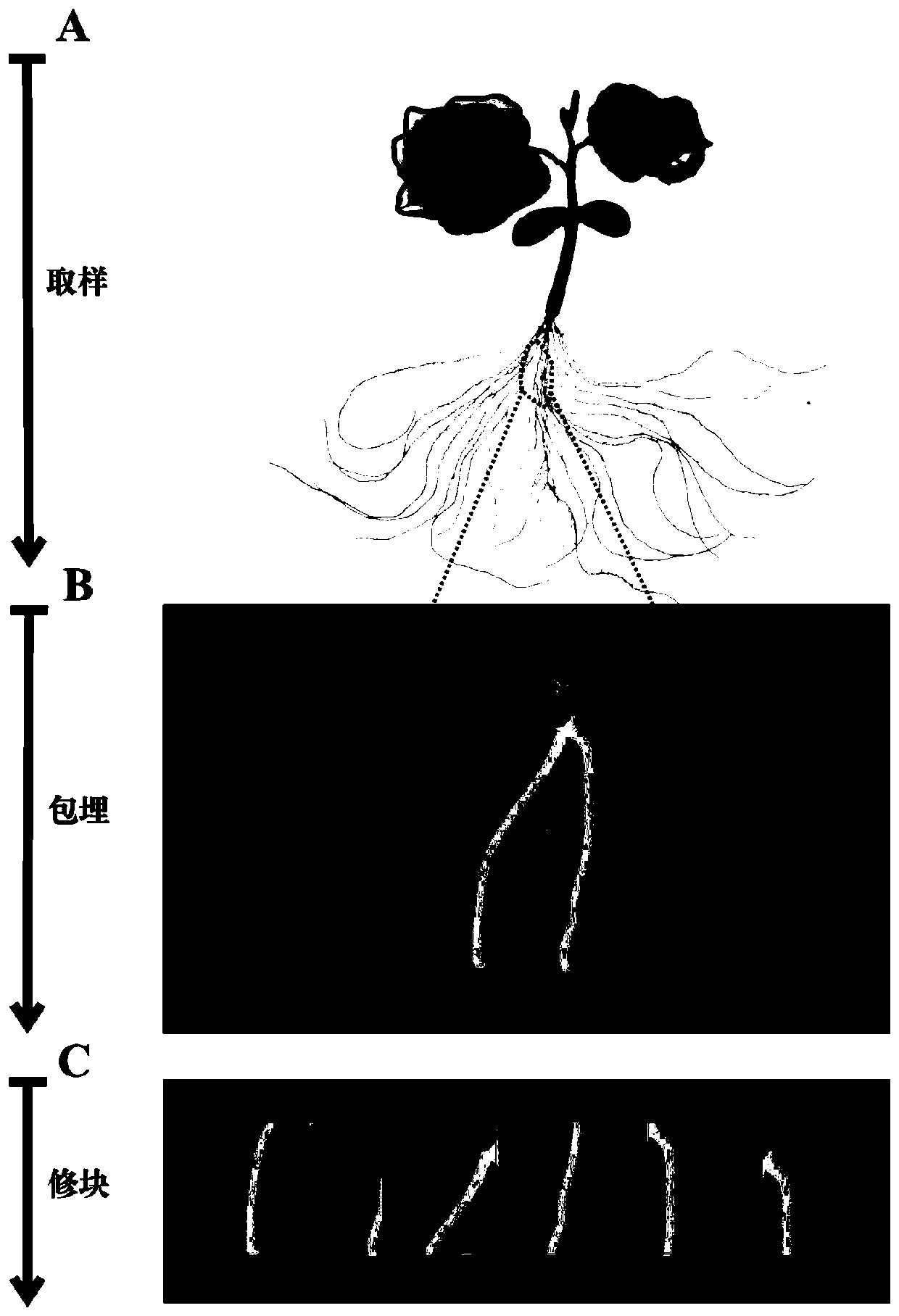
RNA in-situ hybridization optimization method for cucumber seedling gene localization research
ActiveCN110564821AIncreased sensitivityImprove accuracyMicrobiological testing/measurementSlice preparationColor reaction
The invention relates to an RNA in-situ hybridization optimization method for cucumber seedling gene localization research. The method comprises the following steps: firstly, improving a plant culture, sampling and fixing method according to the characteristics of cucumber seedling plant materials so as to obtain a complete plant structure containing target tissues; secondly, optimizing operatingtechniques such as slice preparation, hybridization pretreatment, hybridization incubation slice sealing and chromogenic reaction microscopic examination in a targeted manner so that the reliability,accuracy and specificity of a gene localization result are improved; finally, designing a treatment scheme suitable for cucumber seedling mature tissue after the chromogenic reaction is ended in a targeted mode so that interference of cell inclusions can be eliminated, and directly observing the distribution of hybridization signals. The method is not only used for improving the sensitivity and accuracy of experimental results, but also reduces the economic cost by improving the utilization rate of drugs, reducing the consumption of experimental articles, increasing the yield of reaction products, saving the labor cost and the like, so that the method is a scientific research method considering scientificity, innovativeness and practicability.
Owner:CHINA AGRI UNIV
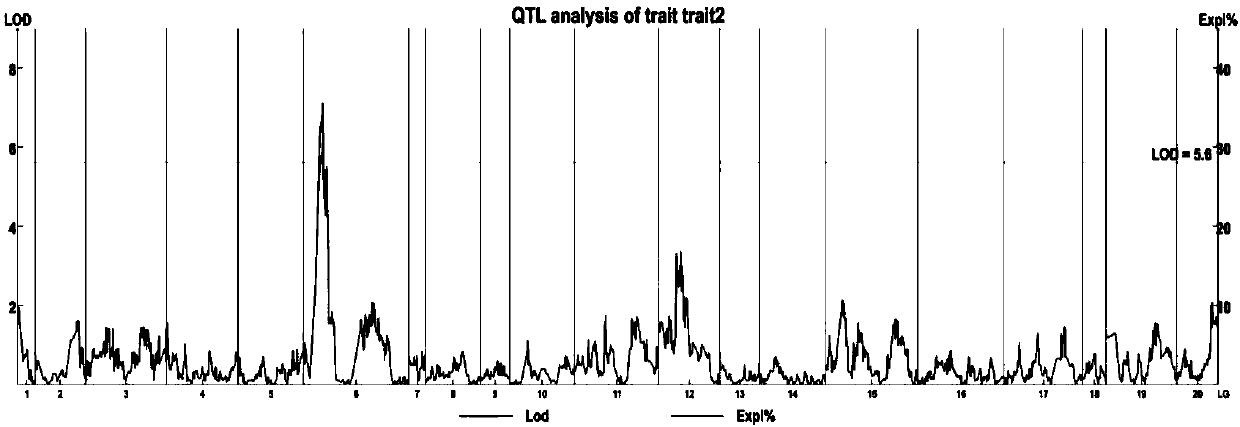
Method for positioning quantitative trait locus (QTL)
InactiveCN101760540AHigh homologyImprove breeding efficiencyMicrobiological testing/measurementQuantitative trait locusComparative genomic analysis
Because quantitative traits present continuous variation and can not be clearly grouped, a quantitative trait locus (QTL) can not positioned by completely utilizing an analyzing method of qualitative trait gene positioning, and a special analyzing method is necessarily developed. A method for positioning the QTL is researched mainly by an analyzing method based on a mark, an analyzing method based on the traits and a comparative genomic analyzing method.
Owner:李祥
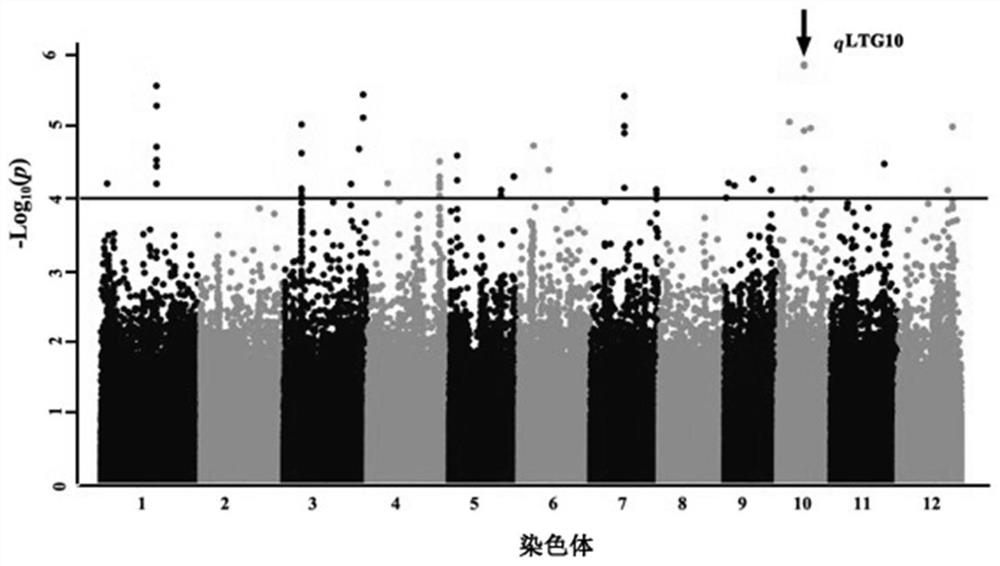
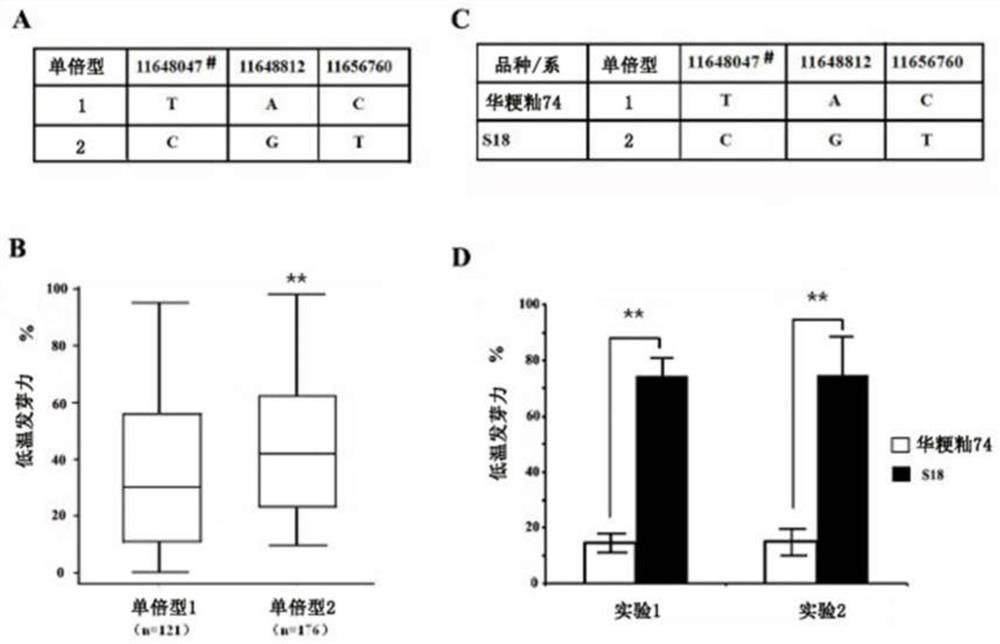
Gene for stably expressing low-temperature germination capacity of rice and molecular marker and application thereof
PendingCN112430603ASkip the phenotyping processSpeed up the breeding processMicrobiological testing/measurementPlant peptidesBiotechnologyHaplotype
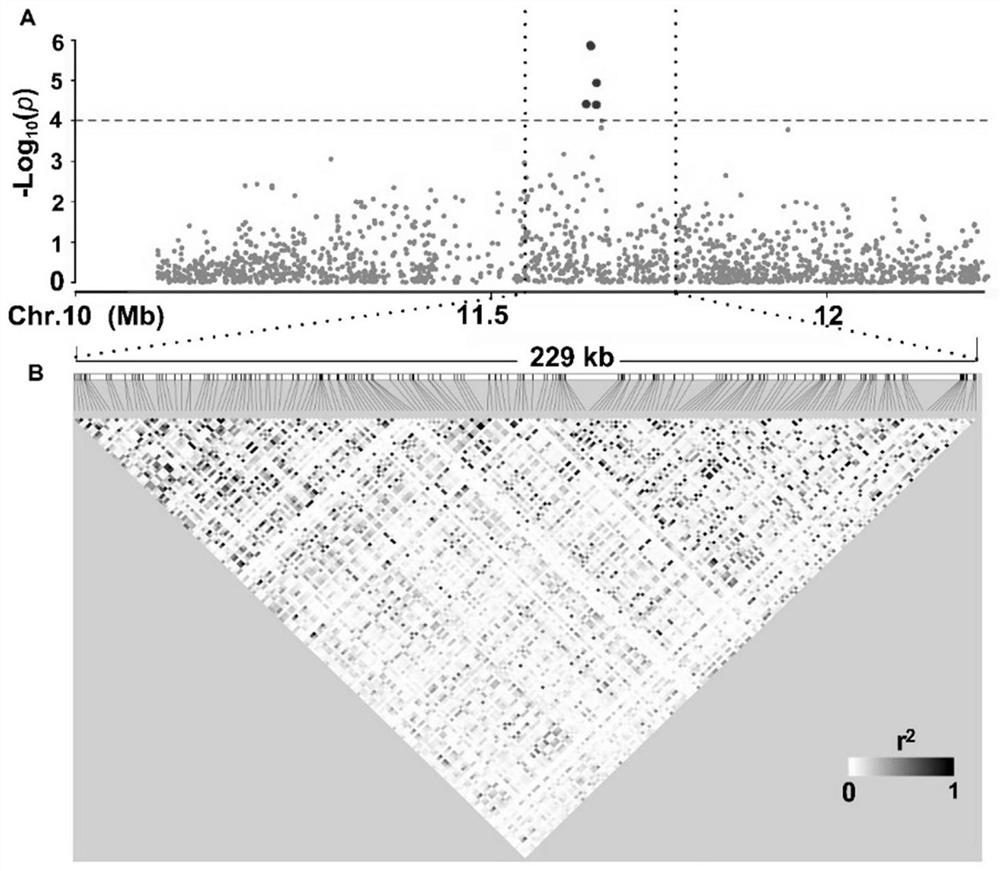
The invention belongs to the technical field of rice trait gene localization, and particularly relates to a rice low-temperature germination capacity stable expression gene and a molecular marker andapplication thereof. The gene is located in the 11551587-11780489bp interval of the tenth chromosome of a rice genome and is named qTLG10, the low-temperature germination capacity of a TAC haplotype is low, the low-temperature germination capacity of a CGT haplotype is high, the molecular marker comprises D22484 or 10ID1, the obtained gene and the molecular marker thereof are beneficial to breeding of rice varieties with strong low-temperature germination capacity, and as the identification of the low-temperature germination capacity of the rice needs specific low-temperature conditions and phenotype selection is difficult, the genotype of the low-temperature germination capacity can be accurately judged by screening the qLTG10 haplotype, the molecular marker provided by the invention performs genotype selection on low-generation breeding populations in the seedling stage, and accelerates the breeding process of low-temperature-resistant rice.
Owner:RICE RES INST GUANGDONG ACADEMY OF AGRI SCI
Central body located gene and its use in medicinal region
InactiveCN1657537APromote proliferationPromote cancerOrganic active ingredientsSugar derivativesTumour tissueDisease
A centrosome located gene cep is disclosed, which is prepared through sequencing, biological information analysis, and gene technique. It can be used as the candidate gene for treating the diseases is hemopoietic system and the generation and transfer of tumor, or as the target point of genetically engineered medicine.
Owner:RADIOLOGY INST ACAD OF MILITARY MEDICINE SCI PLA
Method for gene mapping from chromosome and phenotype data
InactiveUS20050064408A1Microbiological testing/measurementProteomicsGene mappingGenetic linkage disequilibrium
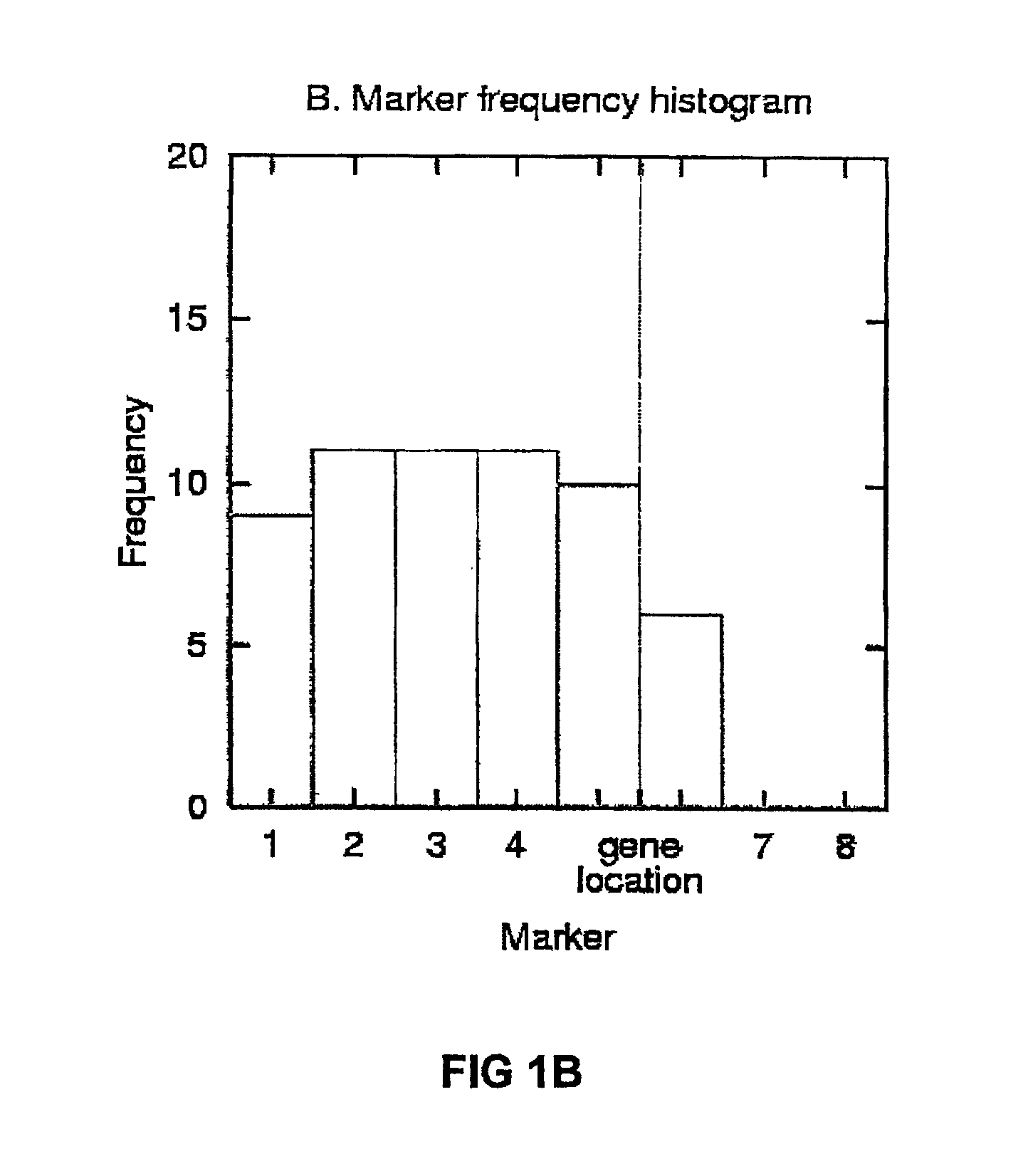
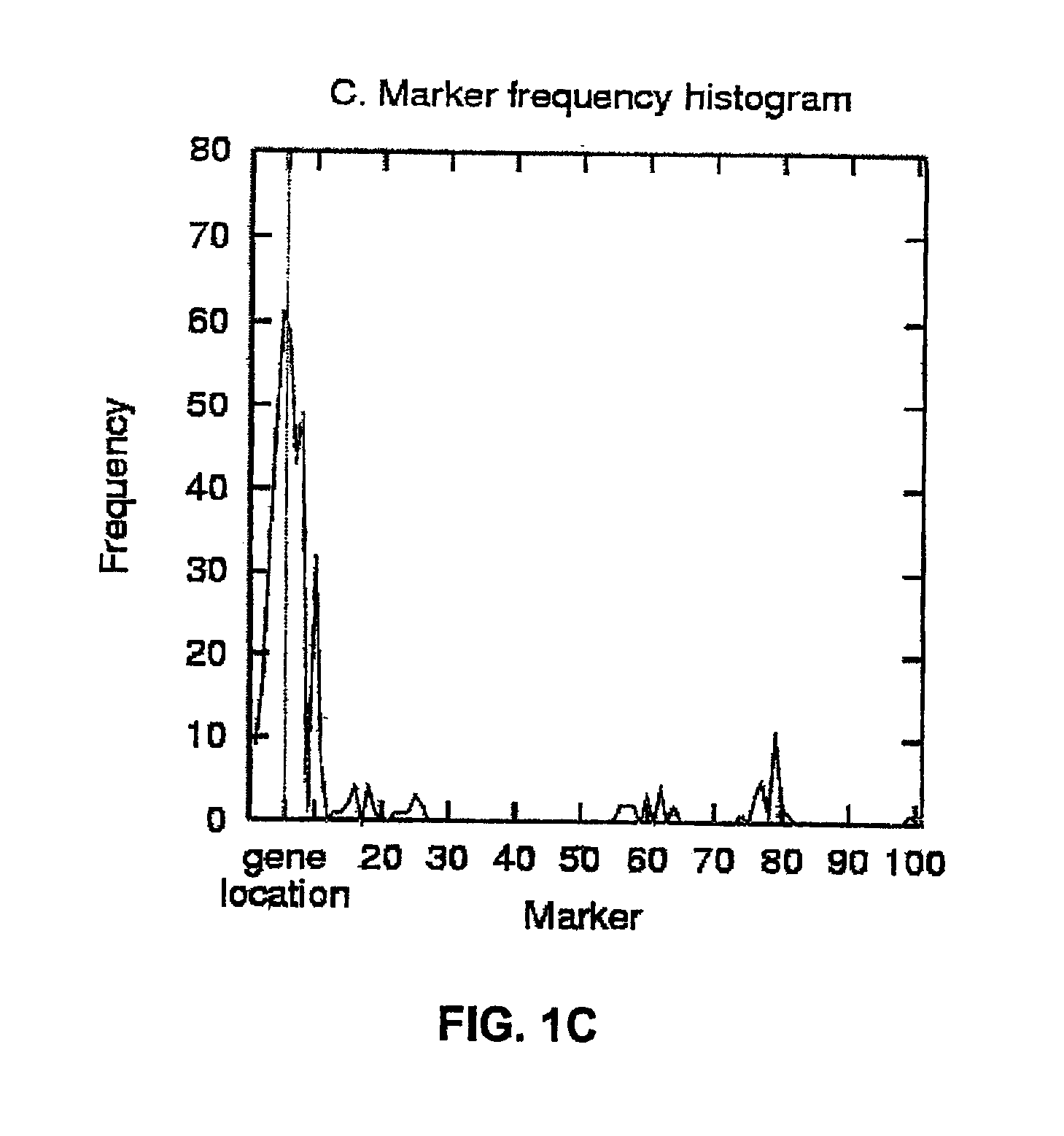
The present invention relates to a method for gene mapping from chromosome and phenotype data, which utilizes linkage disequilibrium between genetic markers mi, which are polymorphic nucleic acid or protein sequences or strings of single-nucleotide polymorphisms deriving from a chromosomal region. The method according to the invention is based on discovering and assessing tree-like patterns in genetic marker data. It extracts, essentially in the form of substrings and prefix trees, information about the historical recombinations in the population. This infor-mation is used to locate fragments potentially inherited from a common diseased founder, and to map the disease gene into the most likely such fragment. The method measures for each chromosomal location the disequilibrium of the prefix tree of marker strings starting from the location, to assess the distribution of disease-associated chromosomes.
Owner:LICENTIA OY
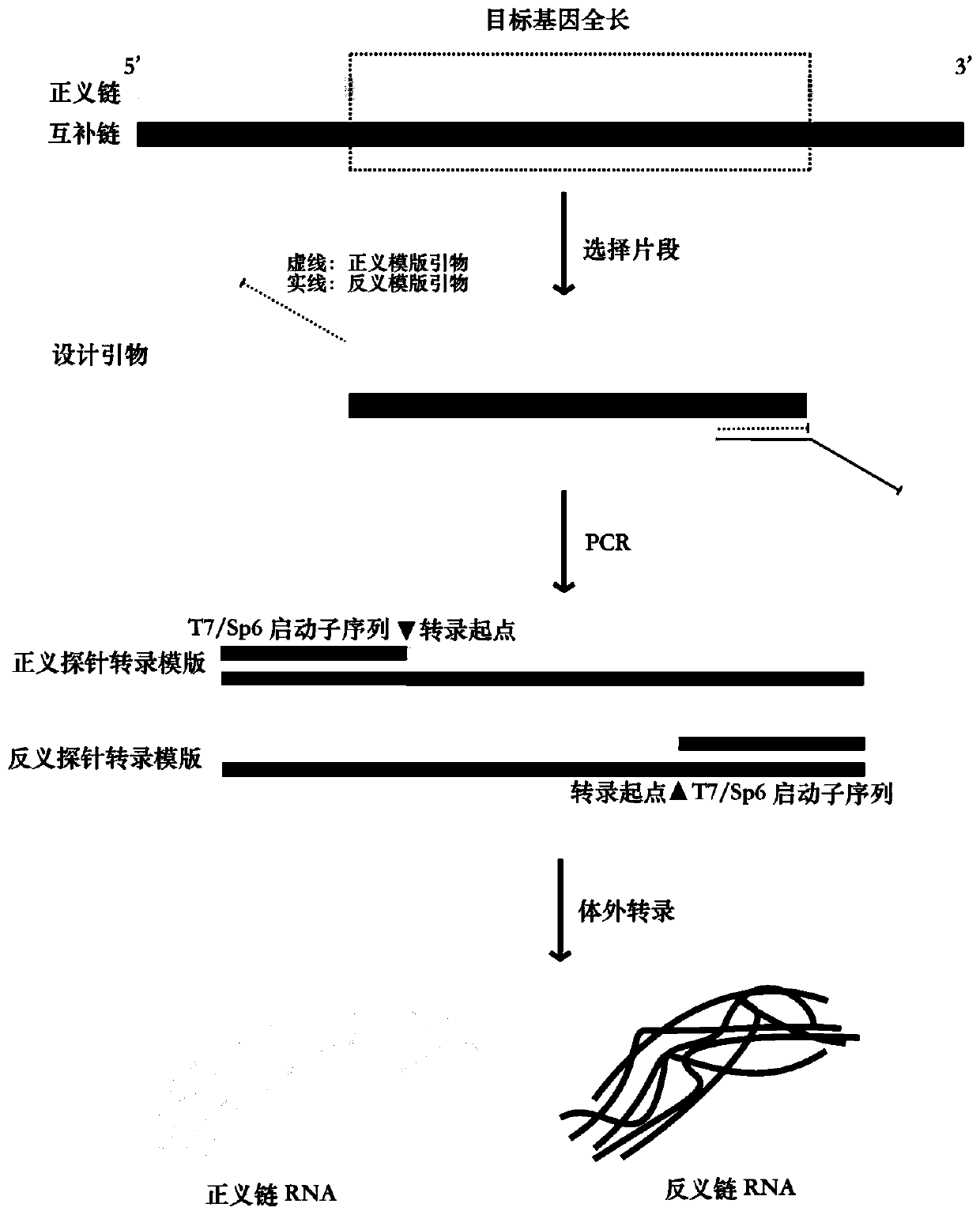
In situ methods for gene mapping and haplotyping
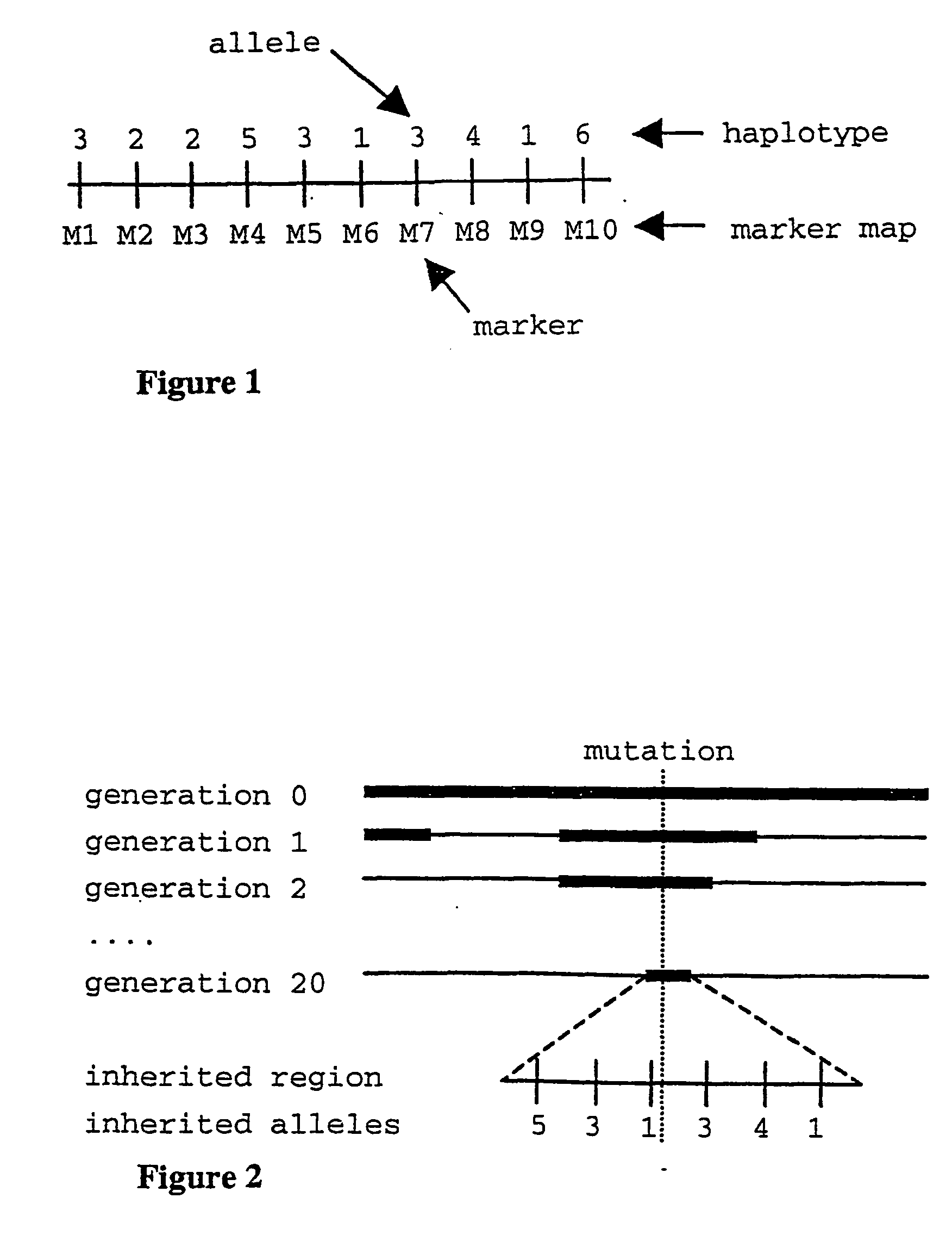
The present invention is directed to in situ methods for providing a definitive haplotype of a subject. The haplotype information generated by the methods described herein is more accurate than that provided by prior art methods that only give an inferred haplotype. Accordingly, in one aspect the present invention provides an in situ method for obtaining genetic information for a polyploid subject, the method including the steps of obtaining a biological sample from the subject, the sample containing: (i) at least one paternally-derived DNA molecule, and / or (ii) at least one maternally-derived DNA molecule, analyzing any one or more of the paternally- or maternally-derived DNA molecules for nucleotide sequence information, wherein the step of analyzing determines whether any two DNA markers are present in cis on one chromosome, or in trans across two sister chromosomes. Use of in situ methods such as FISH allows for the provision of phase-specific information on DNA markers without recourse to methods for physically separating sister chromosomes. Applicants propose that method eliminates the problem of incorrect or misleading inferences concerning the phase of two or more loci within a haplotype, and allows for revelation of two or more participatory genes within a haplotype, uncomplicated by differences in modes of inheritance.
Owner:HAPLOMIC TECH
Maize whole-genome InDel (insertion-deletion) chip and application thereof
ActiveCN110846429AGood effectOptimize detection resultsMicrobiological testing/measurementProteomicsMolecular breedingGene Localization
The invention discloses a maize whole-genome InDel (insertion-deletion) chip and application thereof. By large-scale sequencing mining, InDel sites of maize gene expression regions and whole-genome uniformly distributed InDel sites are determined, and the Affimatrix Axiom based high-flux InDel chip is designed and comprises probes aiming at 1536 InDel sites. The maize whole-genome InDel chip can be used for creating a maize DNA fingerprint database to perform maize variety authenticity identification, maize variety identification or authentication, maize clustering analysis, maize genetic relationship analysis and germplasm resource analysis and can also be used for maize linkage map construction, gene mapping analysis and maize molecular breeding material background selection, and a wideapplication range and remarkable benefits are realized.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Ramie-yield-trait-associated SSR (simple sequence repeat) markers and application thereof
ActiveCN106011261AIncrease productionHigh mapping positioning accuracyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMolecular breeding
The invention discloses ramie-yield-trait-associated SSR (simple sequence repeat) markers which comprise SSR markers RAM200, RAM108 and RAM141. The invention also discloses application of the ramie-yield-trait-associated SSR markers in cultivating ramie new species. The invention also discloses application of the ramie-yield-trait-associated SSR markers in stabilizing or enhancing ramie yield. The SSR markers disclosed by the invention are associated with the ramie stem diameter, and the stem diameter is directly associated with the ramie yield. The SSR markers lay a foundation for high-quality germplasm screening, gene localization and cloning, and molecular marker assisted breeding in future. The SSR markers can be used in ramie molecular breeding and ramie yield enhancement. The SSR markers have important meanings for ramie breeding and production in China in the present stage. Meanwhile, the invention provides beneficial references for molecular marker breeding.
Owner:INST OF BAST FIBER CROPS CHINESE ACADEMY OF AGRI SCI
Molecular marker sisv0832 closely linked to heading date gene in millet
InactiveCN102690818BEasy to set upImprove throughputMicrobiological testing/measurementVector-based foreign material introductionGene mappingGenomic DNA
The invention belongs to the field of molecular biology, relates to a molecular marker and especially relates to a molecular marker SIsv0832 closely linked with a Setaria italica L. Beauv. heading stage gene. The molecular marker SIsv0832 has a sequence shown in the Seq ID NO.1. The invention also relates to primers for amplification of the molecular marker SIsv0832, a use of the molecular marker SIsv0832 and the primers in Setaria italica L. Beauv. heading stage gene mapping or Setaria italica L. Beauv. genetic breeding, and a Setaria italica L. Beauv. breeding method. The molecular marker SIsv0832 links a genomic DNA sequence and the Setaria italica L. Beauv. heading stage gene, and is conducive to establishment of a Setaria italica L. Beauv. molecular marker assistant breeding system. A genetic close linkage distance between the molecular marker SIsv0832 and the Setaria italica L. Beauv. heading stage gene is 3.5cM. In Setaria italica L. Beauv. breeding practices and resource and variety identification, the simple, fast and high throughput utilization of the molecular marker SIsv0832 and the primers for amplification of the molecular marker SIsv0832 can be realized.
Owner:深圳华大基因农业控股有限公司
dCAPS (derived cleaved amplified polymorphic sequence) primer pair for assisting in judging first female node and first male node of pumpkin and application of dCAPS primer pair
ActiveCN108728570AShorten the timeHigh contribution rateMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceThroughput
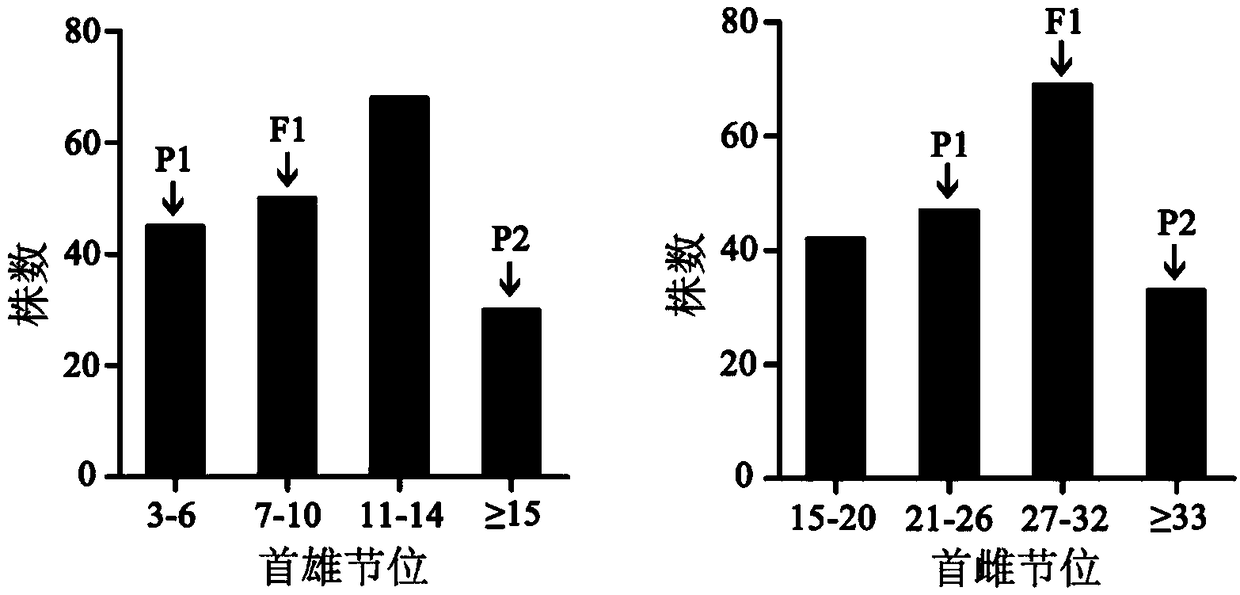
The invention discloses an SNP molecular marker 32622 and a molecular marker 63349 which are closely linked with QTL (quantitative trait loci) of a first female node and a first male node of a pumpkinand application of the molecular markers. The located QTL have a significant incidence relation with the quantitative trait (p is smaller than 0.05), the contribution rate is relatively high, the interpretation probability of the molecular marker 32622 is 26.5%, and the interpretation probability of the molecular marker 63349 is 21.9%; and through the two molecular markers, the heights of the first female node and the first male node of the pumpkin can be respectively predicted. A dCAPS primer pair designed according to the molecular markers can be applied to the improved molecular-assisted breeding of pumpkin varieties in a simple, rapid and high-throughput manner, provides molecular-assisted selection technology support for the realization of the early identification and screening of the traits of the first female node and the first male node of the pumpkin, and greatly shortens the time of traditional gene localization; and after verification, the predicted accuracy rates of the first female node and the first male node of a plant are 77% and 83% respectively.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
Molecular marker SIsv0408 capable of being closely linked to Setaria italica pollen color gene
InactiveCN102154284AImprove throughputEasy to set upMicrobiological testing/measurementVector-based foreign material introductionPollenDNA
The invention belongs to the field of molecular biology and relates to a molecular marker, particularly relating to a molecular marker capable of being closely linked to a Setaria italica pollen color gene. The molecular marker has a sequence as shown in Seq ID No.1. The invention also relates to primers for amplifying the molecular marker, the applications of the molecular marker and the primersin pollen color gene localization of Setaria italica or genetic breeding of Setaria italica, and a Setaria italica breeding method. The molecular marker links the genome DNA sequence with the Setariaitalica pollen color gene and helps the establishment of Setaria italica molecular marker-assisted breeding systems. The distance of genetic close linkage between the molecular marker and the Setariaitalica pollen color gene is 0.3 cM. The molecular marker and the molecular marker amplification primers provided in the invention are designed for the convenient, rapid and high-throughout application in the breeding practices and the resource and variety identification of Setaria italica.
Owner:深圳华大基因农业控股有限公司
Molecular marker SVmc3 closely linked with husked millet color genes
InactiveCN106811508AImprove throughputEasy to set upMicrobiological testing/measurementDNA/RNA fragmentationHigh fluxBiology
The invention belongs to the field of molecular biology, and relates to a molecular marker, in particular to a molecular marker closely linked with husked millet color genes. The molecular marker contains a sequence as shown in SEQ ID NO.1. The invention also relates to a primer pair for extending the molecular marker, a detection method of the molecular marker, an application of the molecular marker or the primer pair in millet assisted breeding or location or detection of the husked millet color genes, a millet assisted breeding method, and a method for screening the molecular marker. The molecular marker provided by the invention combines genome DNA sequence and the husked millet color gene, so that the establishment of the millet molecular marker assisted breeding system is facilitated; and a genetic close linkage distance of the molecular marker and the husked millet color gene is 1.2cM. The molecular marker and the molecular marker extended primer can be conveniently and rapidly used for the millet breeding practice and the resource and variety identification in a high flux manner.
Owner:SHENZHEN BGI AGRI & CYCLE ECONOMIC TECH +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com