Lancea tibetica microsatellite molecular markers
A technology of molecular markers and succulent grass, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, DNA preparation, etc., which can solve the problems of difficult artificial planting and exacerbation of endangered processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: The preparation method of the microsatellite molecular marker of S. sarcocarpa
[0031] 1. Construction of Sarcocarpa DNA library
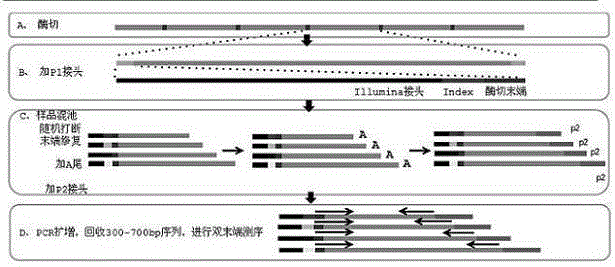
[0032] 1.1 Extraction of genomic DNA: the genome of S. succulents was extracted by the CTAB method, and then digested with restriction endonucleases, and the digested DNA fragments were connected to the adapter p1.
[0033] 1.2 Mixed pool: the interrupted DNA, after end repair, add an A tail to the other end of the interrupted DNA, and then add a p2 linker.
[0034] 1.3 Sequencing: Perform PCR amplification on the above DNA fragments, recover 300-700bp sequences, and perform paired-end sequencing on the Illumina HiSeq sequencing platform after purification. Schematic diagram of DNA library construction and library inspection of S. figure 1 .
[0035] 2. Sequencing data quality control
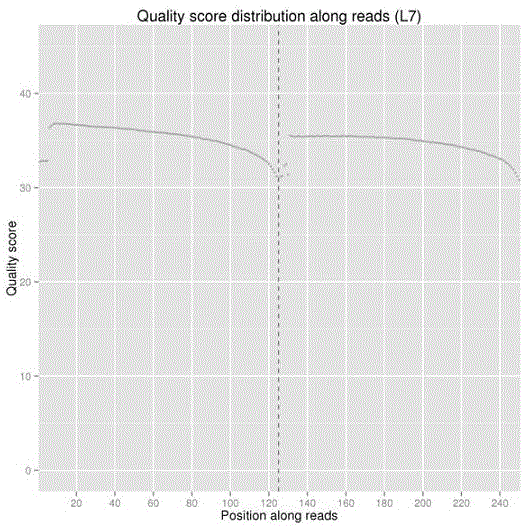
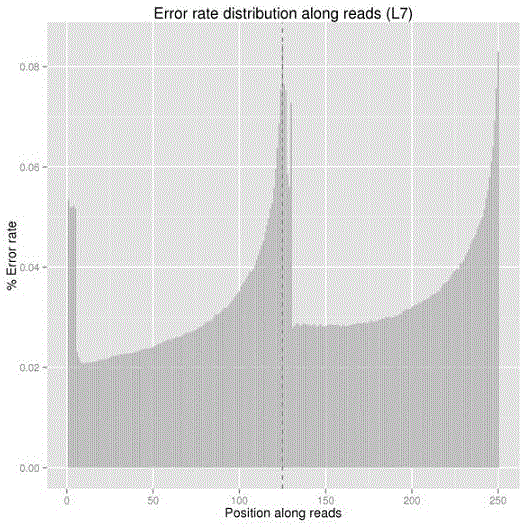
[0036] 2.1 Data quality inspection: Sequence 125bp at each end, use IlluminaCasava1.8 for base calling (BaseCalling), obtain the original se...
Embodiment 2
[0051] Embodiment two: Carry out the genetic diversity analysis of succulent grass with described 10 pairs of primers
[0052] The expected heterozygosity and observed heterozygosity were calculated by genepop software, and the inbreeding coefficient was calculated to describe the characteristics of the 10 microsatellite DNA polymorphisms in S. sarcocarpa. The results are shown in Table 5.
[0053] It can be seen from Table 5 that the 10 microsatellite sequences of the present invention have diversity in 56 individuals of 3 populations. It can be seen that the 10 microsatellite markers of the present invention can be used for genetic diversity analysis, construction of plant genetic map, gene positioning, variety identification, germplasm preservation, analysis of quantitative trait genes, evolution and kinship research, etc. It is characterized by good repeatability and is a reliable and effective molecular marker.
[0054] The results of 10 primers of Table 5 S. succulent ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com