Molecular marker related to dwarfing of corn plant
A molecular marker, corn technology, applied in the field of molecular genetics, to achieve the effect of shortening the breeding cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
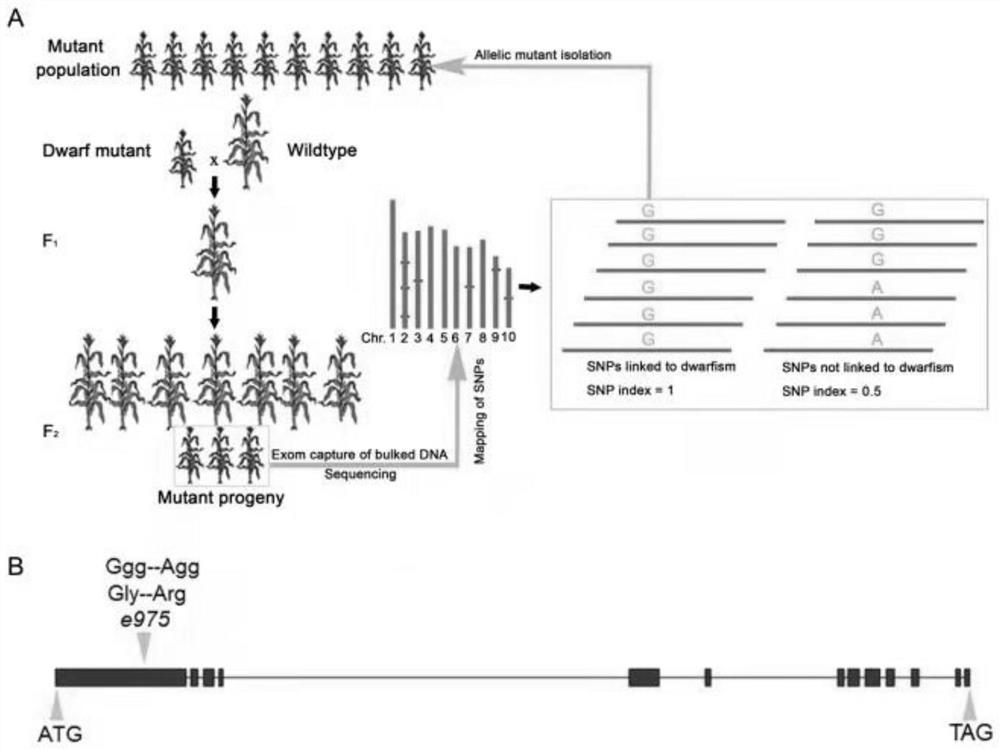
[0027] Example 1: Molecular identification of the e975 mutant
[0028] According to the EcMutMap gene cloning method (principle see figure 1 A), select 35 individual plants with e975 mutant phenotype from the F2 segregation population prepared by wild-type B73 and e975 mutant, and extract DNA from the leaves. Karroten kit (Karroten, sK2304) is used for DNA extraction. For specific steps, refer to manual. Then 500ng of each individual plant was mixed in equal amounts and sent to the company for exon capture sequencing. Taking the B73 genome published online as a reference, GATK (v2.1-9) was used for variation mining. SnpEff v3.6c was used to annotate variant sites. The index (SNP-InDex) of single nucleotide substitutions (SNPs) in the mutation results was calculated using a self-written Perl script. The variation linked to the mutant phenotype is selected according to the following conditions: 1) G→A(C→T); 2) SNP-InDex>=0.9; 3) The variation type is non-synonymous mutation...
Embodiment 2
[0029] Example 2: Molecular marker design of e975 mutation site
[0030] The genome sequence of 250 bp was intercepted on both sides of the mutation site and used as a template for primer synthesis (as shown in SEQ ID NO.1-2, including a total of 501 bp of the mutation site), and Primer Premier 5 was used to design around the 251st base of the template Forward primer and reverse primer. Primer design principles: Tm value is around 60°C, product size is 250-550bp, and primer length is 22-24bp.
Embodiment 3
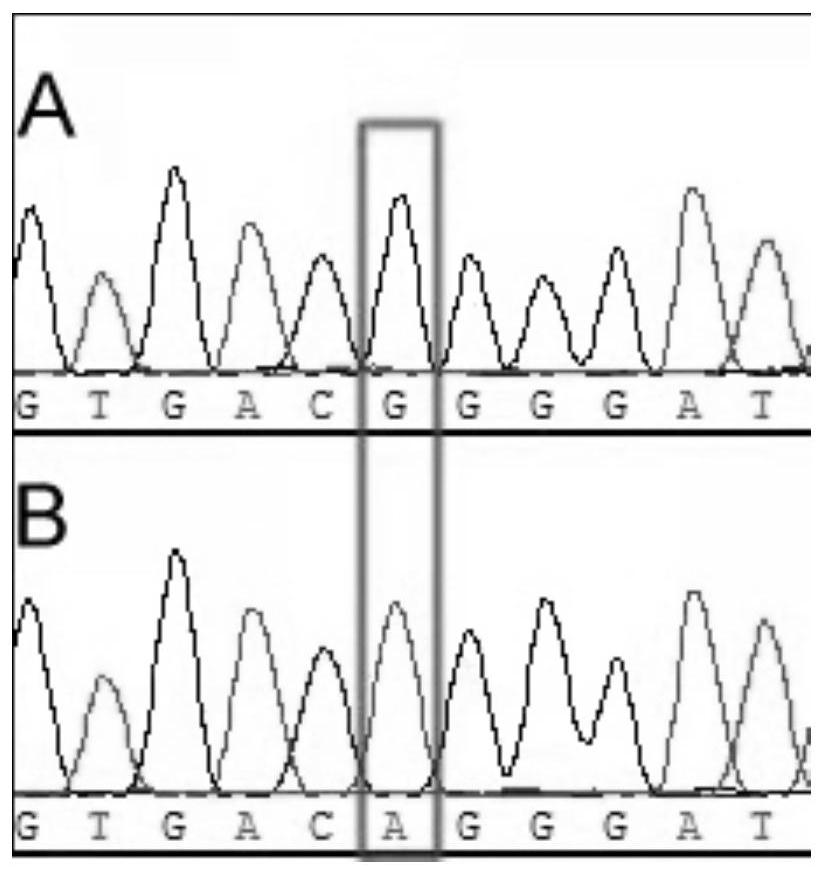
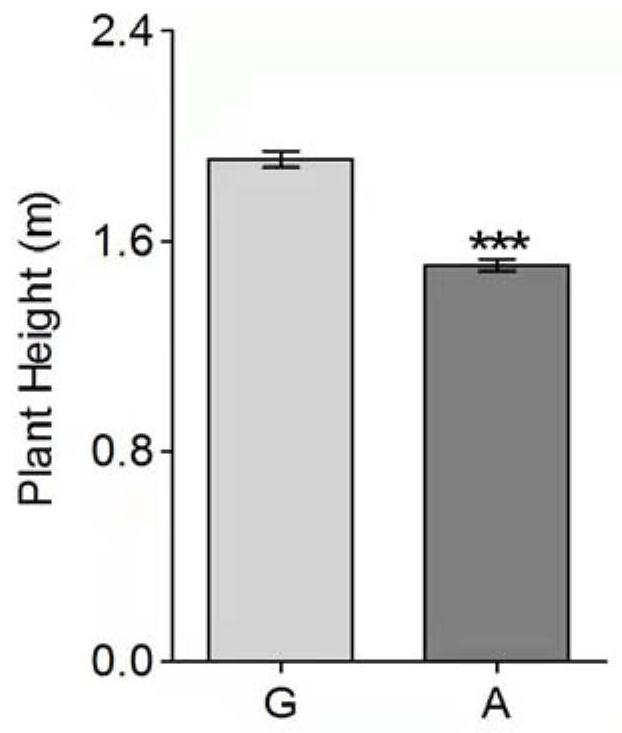
[0031] Example 3: Verification of wild-type and e975 mutant molecular markers
[0032] The primers were synthesized by Nanjing Qingke Biotechnology Co., Ltd., and the specific sequences are as follows.
[0033] Forward primer: 5'-CGATTTGCTGTAGAGGAACTCAGG-3'(SEQ ID NO.3),
[0034] Reverse primer: 5'-AATCAAACTTTCCGCCGAAGGTGT-3' (SEQ ID NO.4).
[0035] The total volume of the PCR system is 25 μL, including 1 μL of forward and reverse primers (10 μmol / L), 1 μL of genomic DNA, 0.2 μL of EasyTaq DNA polymerase, 2.5 μL of buffer, 2 μL of dNTPs, ddH 2 O to make up to 25 μL.
[0036] The PCR reaction program was: pre-deformation at 95°C for 5 min; denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s, a total of 35 cycles; over-extension at 72°C for 5 min; storage at 16°C.
[0037] The length of the PCR amplification product is 298bp, and then it is separated by 1% agarose gel electrophoresis, and the PCR product that meets the target fragment siz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com