Maize whole-genome InDel (insertion-deletion) chip and application thereof
A whole-genome and corn technology, applied in genomics, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problem of expensive chip detection platform, difficult to scale and automate, and can not take into account high-throughput of sites and high-throughput of samples Quantity and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 3
Embodiment 1
[0089] Example 1 Obtaining the InDel polymorphic site of the whole genome of maize and the synthesis of the whole genome chip
[0090] (1) Selection of InDel sites in 40K InDel chips
[0091] (1) Resequence 100 corn materials, each corn material has a sequencing data of about 10Gb, use bwa software (0.7.0) to compare the resequenced sequences of corn materials to the corn reference gene sequence, use Samtools and GATK software Mining small InDel variants were carried out. The length of the InDel sequence ranged from 1 to 20 bp, and a total of 7,843,126 InDel variant sites were excavated.
[0092] (2) In order to make InDel sites have the potential to adapt to electrophoretic analysis, InDel sites with a length of 3-10 bp were selected, and there were 1,705,519 InDel sites in total. These InDel sites were screened using the following conditions:
[0093] a, InDel locus with 2 alleles retained;
[0094] b, filter out InDel sites with MAF values less than 0.1;
[0095] c, r...
Embodiment 2
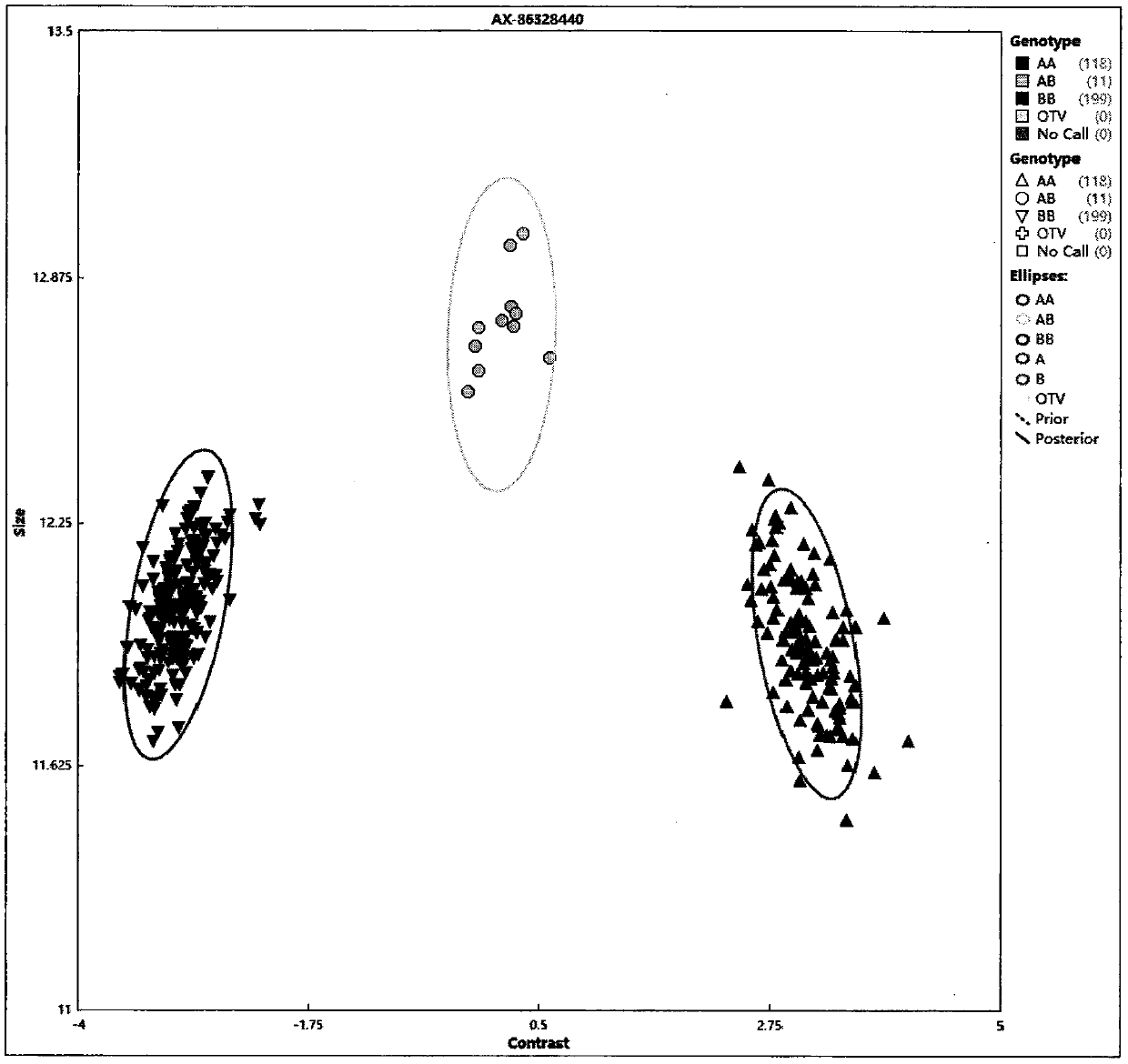
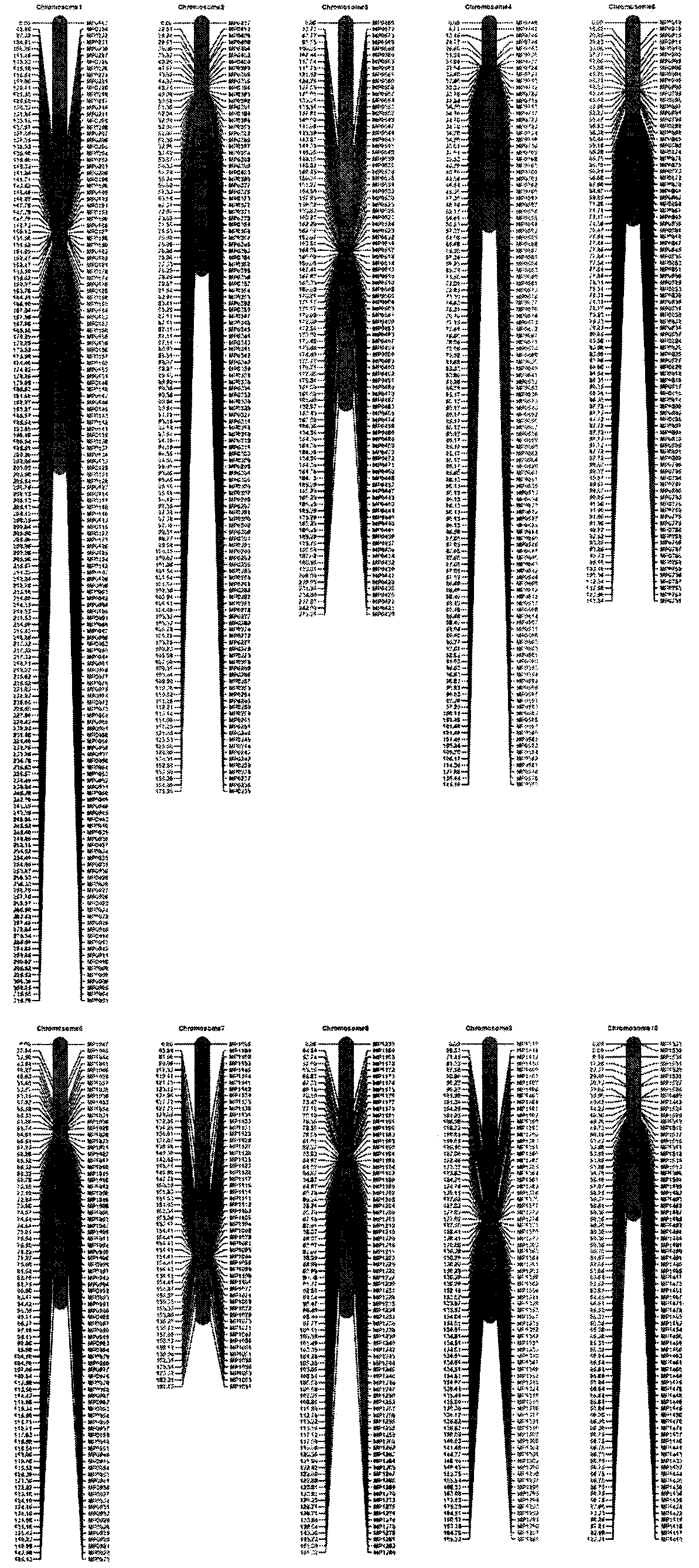
[0105] Example 2 Application of corn 40K InDel chips in the analysis of corn germplasm resources
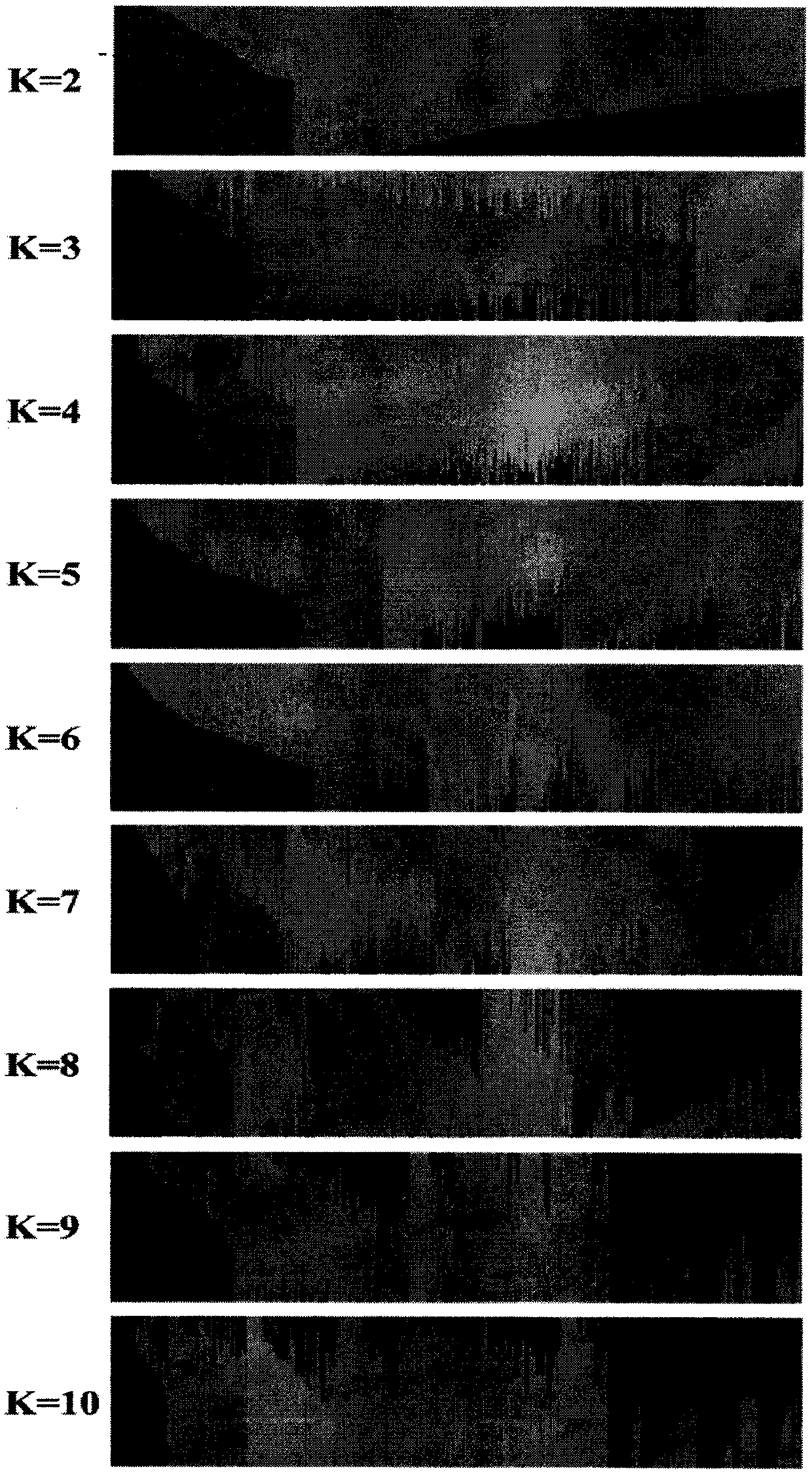
[0106] Using the MaizeIDP40K InDel chip synthesized in Example 1, 335 maize inbred lines from a wide range of sources were used for InDel marker genotyping analysis. The chip detected 335 corn inbred lines. The chip detection results passed the quality control test of the data analysis software Axiom analysis suite, and the sample pass rate was 99%. The missing rate and MAF value of the genotyping data of each InDel marker were calculated, and the data were filtered out. 24968 high-quality InDel markers ( figure 1 ), accounting for 61.2%. Using FAST STRUCTURE to analyze the population genetic structure of 335 maize inbred lines using the genotyping data of InDel markers. Population genetic structure analysis shows that when K=10, the ΔK value is the largest, that is, the analyzed inbred line population of the present invention can be divided into 10 groups, which are respective...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com