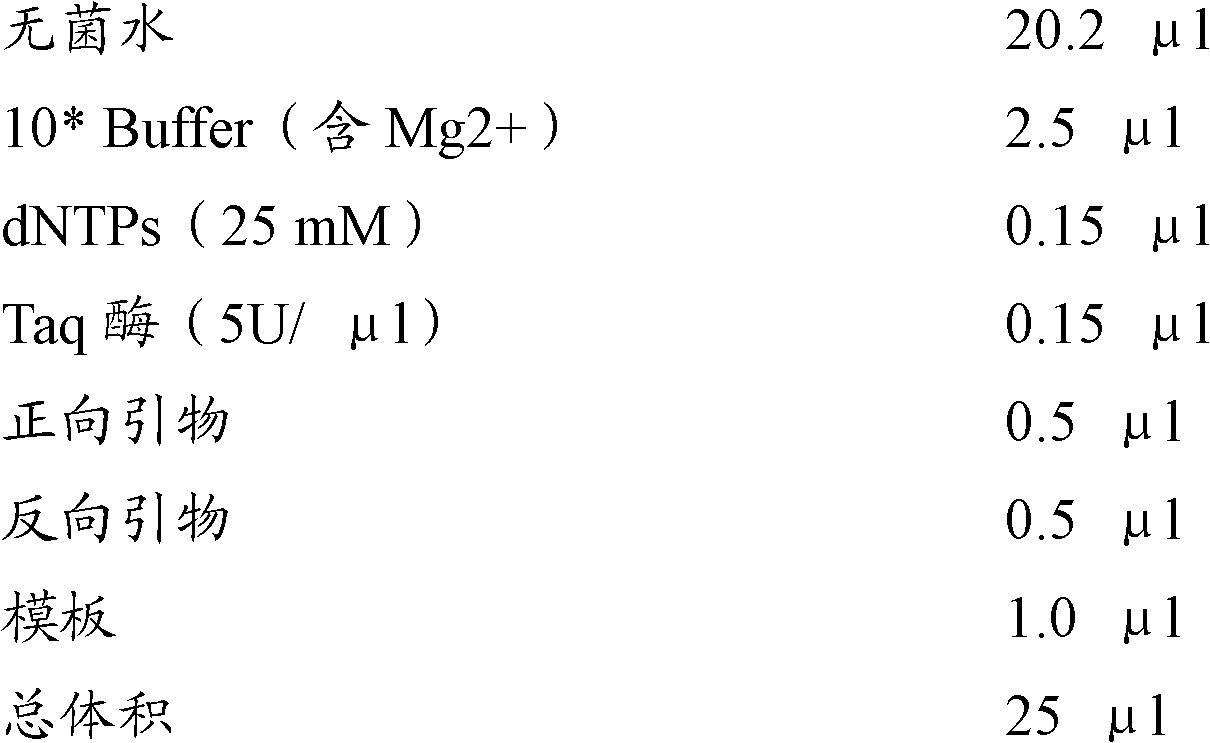Molecular marker SIsv0408 capable of being closely linked to Setaria italica pollen color gene
A technology of molecular markers and color genes, applied in the field of molecular biology, can solve problems such as no literature reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Construction of millet F2 generation segregation population
[0040] Male parent: Kangnabujing, high plant type, long and narrow flag leaves, red bristles, red glumes, fertile, greenish leaves, yellow-white pollen, late heading stage. The male parent is Zhang Gu No. 1 seed.
[0041] Female parent: Not resistant to Nabujing, short plant type, short and wide flag leaves, green setae, green glumes, partly sterile, yellowish leaf color, brown pollen, early heading stage. The female parent is the seed of millet A2 male sterile line.
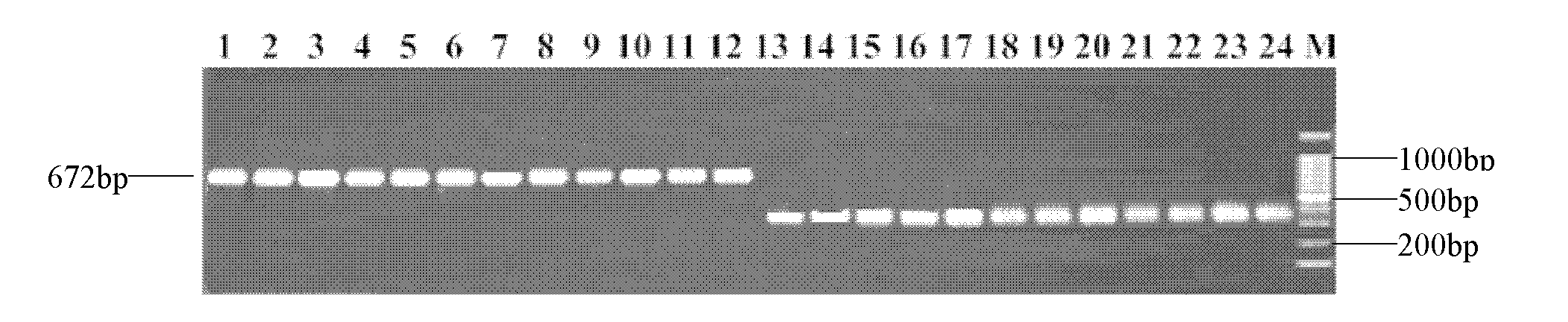
[0042] F2 population construction: the male parent and the female parent were crossed to obtain the F1 generation (F1 pollen color is orange), and F1 was self-crossed to obtain F2. Among them, F1 is the No. 3 seed of Zhang Zagu. A total of 480 F2 plants were obtained, of which 120 were yellow-white pollen, 240 were orange, and 120 were brown.
[0043] See the Chinese patent application "Molecular marker SIsv0372 closely linked to...
Embodiment 2
[0044] Example 2: Extraction of parental and F1 generation, F2 generation individual genomic DNA
[0045] Genomic DNA of the parents, the F1 generation, and 480 F2 generation individuals in Example 1 were extracted by the CTAB method, and the specific methods were as follows:
[0046] (1) Weigh 1.0g of fresh leaves, cut them into pieces and put them in a mortar, grind them with liquid nitrogen, add 3mL 1.5×CTAB, grind them into a homogenate and transfer them to a 15mL centrifuge tube, then add 1mL 1.5×CTAB into the mortar Rinse and transfer to a centrifuge tube. After mixing, place in a water bath at 65°C for 30 minutes, and shake slowly from time to time during this period.
[0047] Among them, the formula of 1.5×CTAB is as follows (1L):
[0048]
[0049] Add deionized water to make up to 1 L, and add mercaptoethanol with a final concentration of 0.2% (2 ml) before use.
[0050] (2) After cooling to room temperature, an equal volume of chloroform / isoamyl alcohol (24:1) ...
Embodiment 3
[0055] Example 3: Preparation of Molecular Markers
[0056] Using the genomic DNA of the male parent, the F1 generation, or the F2 generation extracted in Example 2 as a template, PCR amplification was performed with a pair of molecular marker amplification primers (Seq ID No.2 and Seq ID No.3).
[0057] The PCR reaction system is as follows:
[0058]
[0059] The PCR reaction procedure is as follows:
[0060] Pre-denaturation at 94°C for 5 minutes; denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 40 seconds, and 35 cycles; final extension at 72°C for 3 minutes. PCR amplification products can be stored at 4°C.
[0061] Molecular markers are obtained through the above amplification process, and the amplification product is preferably purified after amplification. Sequenced after purification, the result is shown in Seq ID No.1.
[0062] Those skilled in the art can understand that the molecular marker can also be obtained by DN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com