Fluorescent labeled X-STR gene locus multiplex PCR method and application thereof
A fluorescent labeling and locus technology, applied in the field of genetic labeling, can solve the problems such as the inability to use paternity testing and personal identification of the Han population, the need for 3 to 4 weeks to arrive, and the limitation of wide application, and reach the scope of applicability of the test materials. Wide range of inspection materials and reagent consumables, high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Children A (8 years old) and B (6 years old) accepted by the same orphanage are suspected to be full siblings and requested a paternity test.
[0038] experiment procedure:
[0039] 1. Collect hairs with follicles from A and B, clip 3 hair follicles and use Chelex-100 to quickly extract DNA, add 100 μL 5% Chelex-100 after centrifugation, incubate at 56°C for 30 minutes, incubate at 98°C for 10 minutes, 10000 rpm After centrifugation for 2 min, the supernatant was aspirated for PCR amplification.
[0040] 5×PCR Reaction mix 2 μL 10×STR Primer mixture 1 μL Hs Taq DNA polymerase (5 U / μl) 0.2 μL DNA template 1 μL Sterilized ultrapure water 5.8 μL
[0041] 5×PCR reaction mixture containing dNTP 200 mmol / L, MgCl 2 1.5mmol / L.
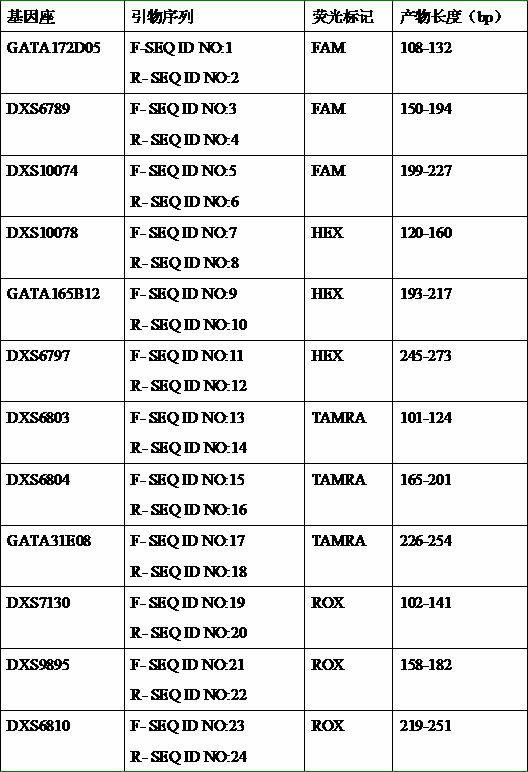
[0042] In the above primer mixture, the primer sequences and their fluorescent labels for each locus are shown in Table 1.
[0043] Table 1 Gene loci and their primer sequences and fluorescent marker...
Embodiment 2
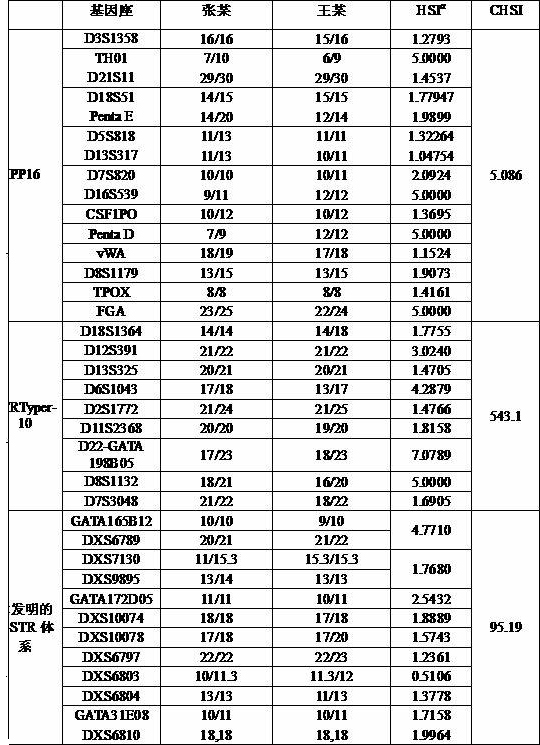
[0058] The X-STR fluorescence-labeled multiplex PCR method was used to identify whether the two daughters Zhang and Wang, whose parents both died, were half-sisters of the same father.
[0059] experiment procedure:
[0060] 1. Collect oral swab samples of Zhang and Wang, extract DNA by Chelex-100 rapid extraction method, add 200 μL 5% Chele-100 after centrifugation, incubate at 56°C for 30 minutes, incubate at 98°C for 10 minutes, and centrifuge at 10,000 rpm2 min, the supernatant was aspirated for PCR amplification.
[0061] 2. PCR amplification: with embodiment 1.
[0062] 3. Capillary electrophoresis: the same as in Example 1.
[0063] 4. Genotyping: Same as Example 1.
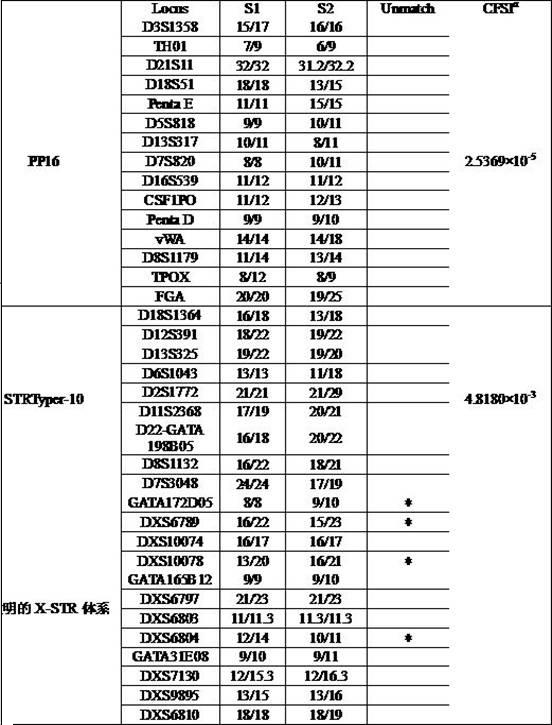
[0064] 5. Simultaneously, 24 autosomal STRs were also typed using the autosomal STR multiplex amplification system PowerPlex® 16 kit (abbreviated as PP16) and STRTyper-10 kit (abbreviated as STRTyper-10). Typing methods were carried out according to the respective manuals.
[0065] Result analysis: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com