Physalis angulata cpSSR-labeled primer developed based on chloroplast genome sequence and its application
A technology of chloroplast genome and marker primers, applied in the field of plant biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Example 1 Acquisition of the chloroplast genome sequence
[0097](1) Extraction of total genomic DNA. Cut 100 mg of fresh leaves of the live sample of Kuanthus, quickly grind it into a powder with liquid nitrogen, quickly transfer the powder to a 1.5mL centrifuge tube, and use the CTAB method to extract the total genomic DNA. The integrity of the total DNA in the sample was detected by electrophoresis using 1.0% agarose gel, and the concentration was detected by an ultraviolet spectrophotometer.
[0098] (2) Long PCR amplification. The total DNA of the extracted samples was amplified by Long-range PCR amplification technology. The amplification products were mixed to a total DNA concentration of ~6 μg / L. The DNA fragments were interrupted by physical methods, and the Illumina sequencing library was constructed according to the technical guidelines of Illumina, and the size of the library fragments was 500bp. Then the sample was loaded and sequenced on the Illumina M...
Embodiment 2
[0101] Example 2 Identification of cpSSR sites and design of cpSSR primers
[0102] (1) cpSSR site screening
[0103] Use the MISA perl script software (http: / / pgrc.ipk-gatersleben.de / misa / ) to search for the SSR sites distributed in the chloroplast genome of Sophora japonica, and the parameter settings are: mononucleotide repeat sequence (mononucleotide), repeat Unit ≥ 10; dinucleotide repeat sequence (dinucleotides), repeat unit ≥ 5; trinucleotide repeat sequence (trinucleotide), repeat unit ≥ 4; four (tetranucleotide), five (pentanucleotide), six (hexanucleotide) nucleosides Acid repeat sequence, repeat unit ≥3.
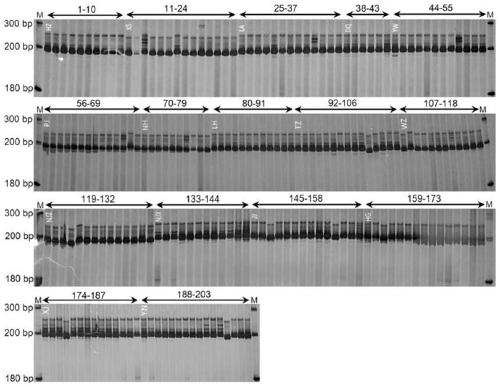
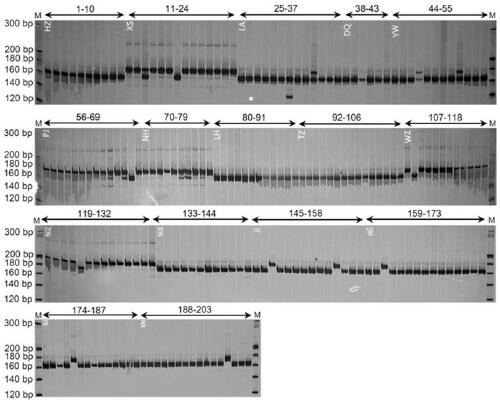
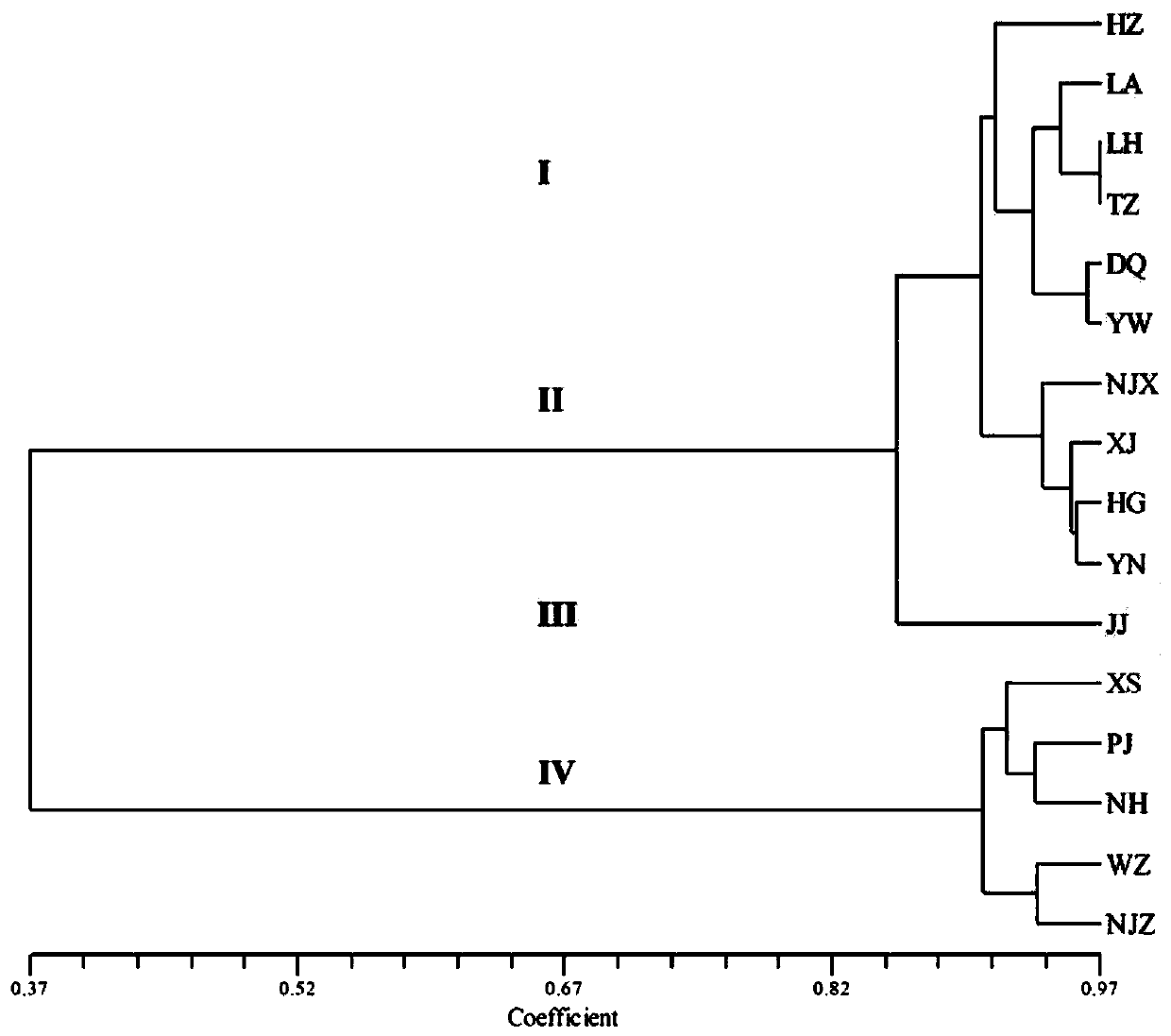
[0104] Through the screening of cpSSR sites, a total of 57 SSR sites were detected from the genome sequence (see Table 1), of which there were 39 single-base repeats (14 A bases, 1 C base, and 24 T bases) ); there are 6 dibasic repeats (3 AT repeats, 3 TA repeats); 5 tribasic repeats, and 7 tetrabasic repeats. The number of A and T repeats accounted for about 6...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com