Linker for detecting low-frequency variation, linker mixture and corresponding method
A mixture and variation technology, applied in the field of biological information, can solve the problems affecting the application and promotion of double-stranded molecular tag technology, the difficulty of accurate quantification of the final linker concentration, and the low success rate of linker preparation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
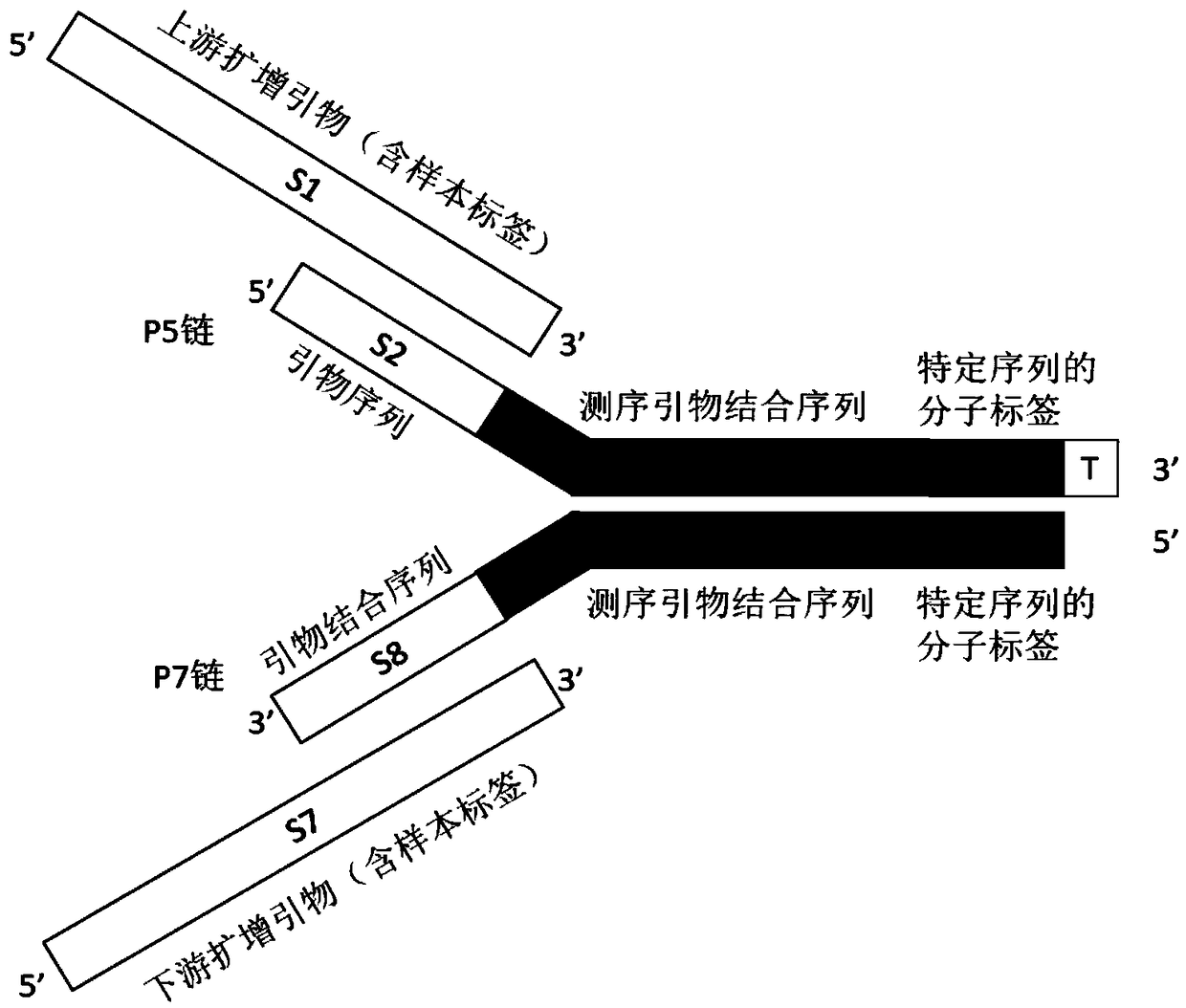
[0098] Example 1 Preparation of adapters containing 7bp+8bp double-stranded molecular tags
[0099] Design the sequences of the linker P5 strand and P7 strand with double-strand molecular tags: the linker mixture is designed to contain 16 kinds of molecular tags, as shown in Table 1 below, 7 pairs of 7 nucleotides in length among the 16 kinds of molecular tags, The remaining 9 pairs are 8 nucleotides in length, and the lengths of the two are staggered by 1 bit. In the first 7 nucleotide sequences of the molecular tag combination, the base ratio of the same vertical position is A:G:T:C=1:1:1:1, and there are at least 3 nucleotide sequences between the molecular tags difference.
[0100] Table 1
[0101]
[0102]
[0103] Centrifuge the synthesized tube at 12,000g for 1 minute to shake the dry powder to the bottom, carefully open the tube cap, dilute the dry powder to 250 μM with LowTE Buffer (10mM Tris-HCl (pH 8.0), 0.1mM EDTA), shake and mix well and place at 4°C Refr...
Embodiment 2
[0114] Example 2 Detection of Low Frequency Somatic Variations in Standards and Tumor Blood Samples
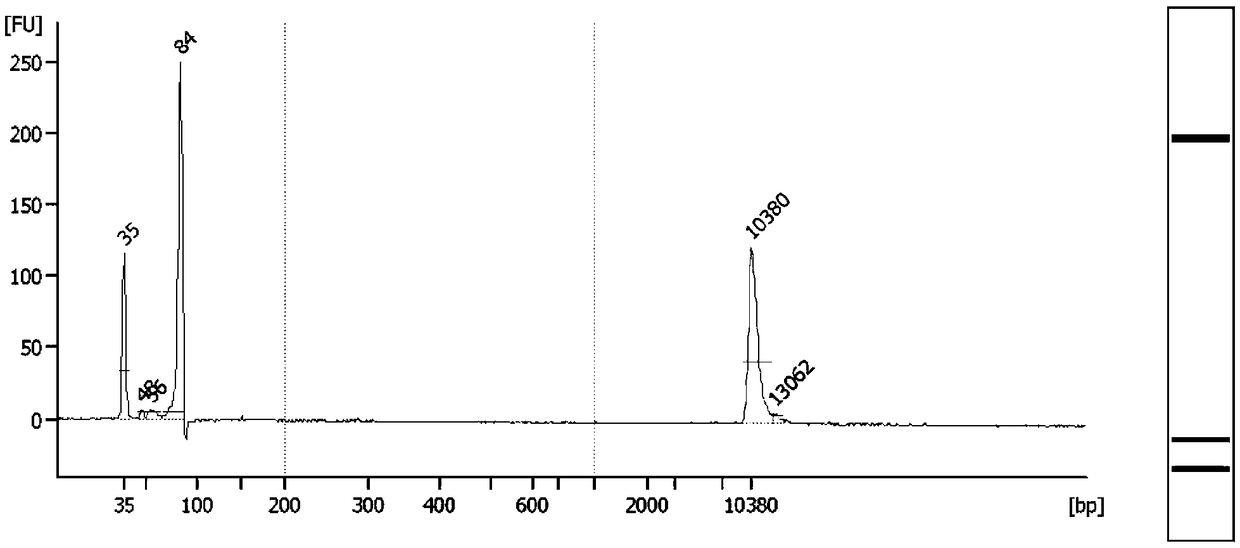
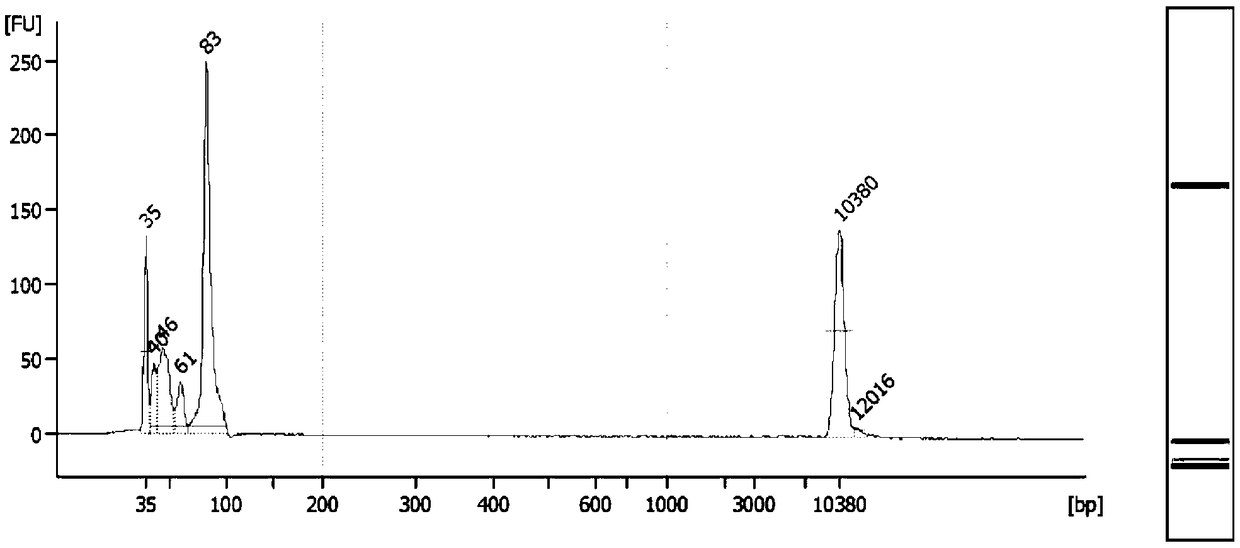
[0115] Preparation of standard DNA: The normal cell line DNA NA18536 was used to dilute the Horizon Discovery standard HD701 and HD753 by different multiples. See Table 4 and Table 5 for the mixture of different dilution multiples and the expected variation frequency. The DNA mixture was sonicated with Covaris S220 until the main peak was around 170bp, which was similar in size to the main peak of cfDNA.
[0116] Table 4
[0117]
[0118]
[0119] table 5
[0120]
[0121] Preparation of cfDNA: Plasma isolated from whole blood of patients was extracted with QIAamp Circulating Nucleic Acid Kit (Qiagen).
[0122] Pre-library preparation: Take KAPA Hyper Prep Kit (Roche) as an example, take 33ng of plasma cfDNA and standard DNA after fragmentation, use KAPA Hyper Prep Kit (Roche) for pre-library preparation, fill in the tail and connect to the example For the linker p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com