Gene modified novel BDFP fluorescent protein and fused protein thereof
A fluorescent protein and protein technology, applied in the field of fluorescent markers, can solve the problems of single emission wavelength and cannot be used in effective combination, and achieve the effect of improving effective brightness, improving effective brightness and stable performance.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
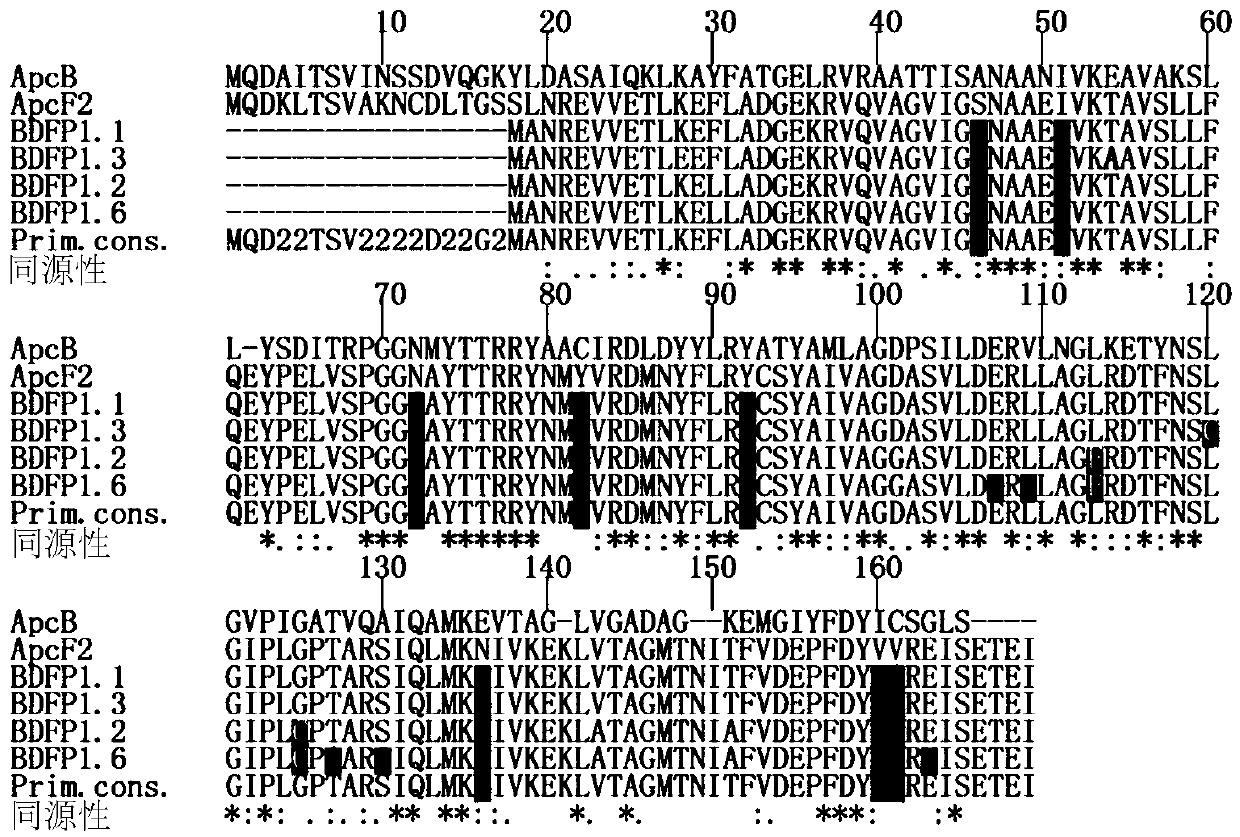
[0073] Example 1. Homologous sequence comparison analysis
[0074] figure 1 A comparison of several homologous sequences is shown, in which ApcF2 and its derivative sequence BDFP1.1 are all from Chroococcidiopsis thermalis PCC 7203; ApcB is from Spirulina platensis, which is not suitable for the growth environment of far-red light and near-infrared light. from figure 1 It can be seen that L113 is highly conserved among ApcB, ApcF2 and BDFP1.1.
[0075] On the basis of the BDFP1.1 sequence (SEQ ID NO.: 2), L113 was subjected to site-directed saturation mutagenesis; the screened mutants were subjected to continuous random mutation through error-prone PCR reactions to establish a mutant library. The mutants in the mutant library were screened by using the method of in vitro spectral property detection and live cell imaging according to the following screening criteria: Compared with BDFP1. 660nm; simultaneously with known fluorescent protein marker iRFP670 (ε·Φ fl =10.2mM -1...
Embodiment 2
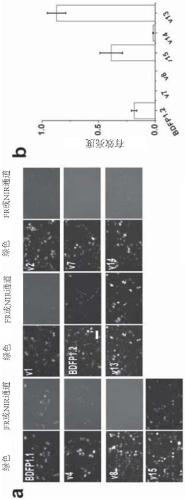
[0082] Example 2. Detection of the effective brightness of each BDFP fluorescent protein in HEK 293T cells
[0083] The nucleic acid expressing each mutant (FPs: IRES: eGFP) was constructed in the expression vector pcDNA3.1, and then transiently transfected into HEK 293T cells, and the fluorescence brightness was observed using an inverted fluorescence microscope. The results are shown in figure 2 middle.
[0084] figure 2 a shows the fluorescence images observed under the green channel and FR / NIR channel, respectively. Because BDFP1.1, v1, v2, v3, v4, v7, v8, v14 in FR / NIR channel, 5s exposure time did not collect images with effective brightness, so take 30s exposure time. The exposure time of other mutants was 5s. figure 2 Quantification of the effective luminance of mutants v6 (BDFP1.2), v7 and v8 is shown in b.
[0085] From the above results, it can be seen that the mutants v1, v2, v4, v6(BDFP1.2), v7 and V8 can all observe fluorescence under the FR / NIR channel. ...
Embodiment 3
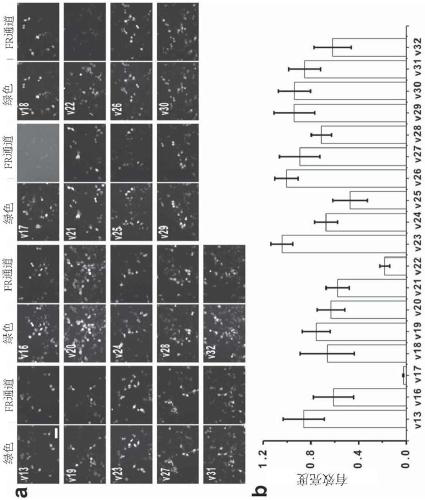
[0086] Example 3. Further Mutant Screening
[0087] In order to further search for mutants with excellent properties, the inventors conducted another round of mutations. With the same screening criteria as in Example 1, 20 mutants v13-v32 were screened out, as shown in Table 3 below.
[0088] At the same time, the spectral properties of the mutants v13-v32 were detected, and the results are shown in Table 4 above.
[0089] From the results in Table 4, it can be seen that the maximum emission wavelength of v13~v32 is similar to that of v6 (BDFP1.2), both are around 670nm, and the molecular brightness is also higher.
[0090]
[0091]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com