An image sample upsampling method based on convolutional self-coding
A technology of convolutional self-encoding and image samples, which is applied in the field of image sample upsampling based on convolutional self-encoding, can solve problems such as large noise, imbalance, and lack of physical meaning in imagery, and achieve simple network scale and training process Simple, visibility-enhancing effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
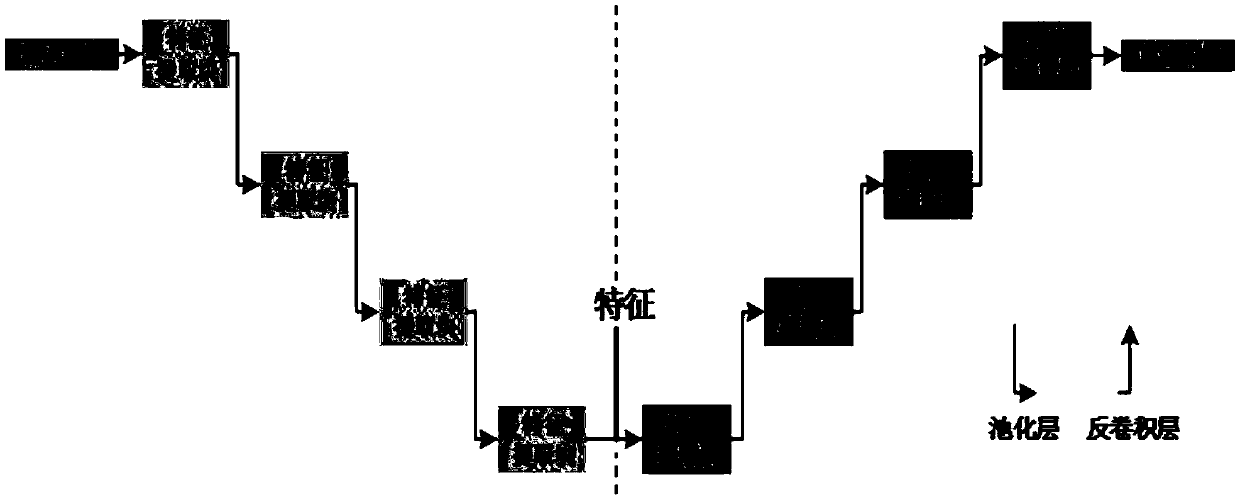
[0038] The embodiment of the present invention proposes an image upsampling method based on convolutional self-encoding, see figure 1 , figure 2 , the method includes the following steps:
[0039] 101: Cut out each 3D MRI sample, cut out the 2D image of the tumor area, and normalize the scale of all 2D images; build a network structure in the form of a cascaded encoder and decoder, and as a model;
[0040] 102: Train the model by setting the learning rate and loss function; use the adaptive moment estimation optimizer to optimize the trained model;
[0041] 103: Input any random positive samples into the trained network, obtain the low-dimensional features extracted by the encoder, calculate the Euclidean distance center point of 8 groups of features, and randomly select 1 group of features among the 8 groups of features to obtain new features;
[0042] 104: Input the new feature into the decoder for image reconstruction, and output the positive sample image.
[0043] Whe...
Embodiment 2
[0052] The following combined with specific examples, image 3 The scheme in Example 1 is further introduced, see the following description for details:
[0053] (1) Minority class sample division, the method is as follows:
[0054] Step 1: Divide the minority class samples into different sets by class.
[0055] Step 2: Randomly select several cases in the positive samples, and randomly select several cases in the negative samples as the verification set; the rest of the data is used as the training set.
[0056] Step 3: Cut out each 3D MRI (magnetic resonance imaging) sample, cut out a 2D image of the area where the tumor is located, and normalize all the 2D images to 224×224.
[0057](2) Network structure construction, the method is as follows:
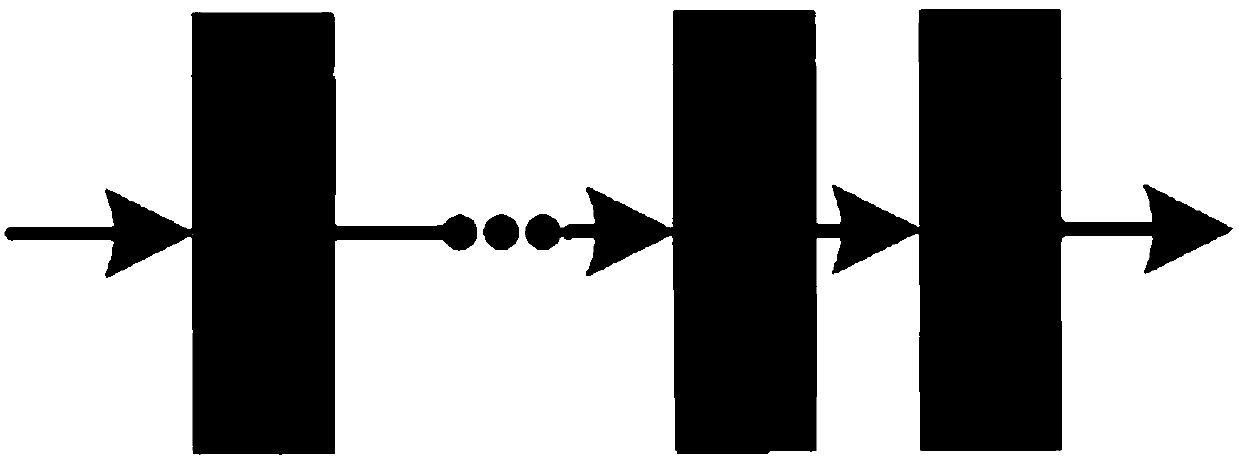
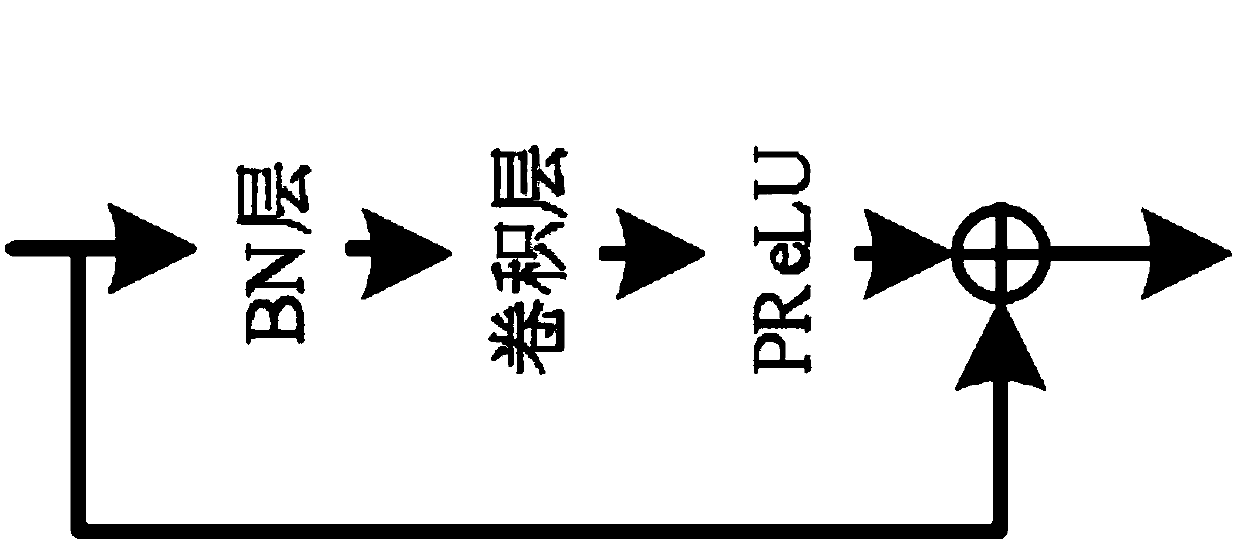
[0058] Step 1: Input 224×224 minority class samples (i.e., positive samples) into the convolutional encoder, which consists of multiple cascaded feature extraction modules and pooling layers. The feature extraction module consis...
Embodiment 3
[0075] The scheme in embodiment 1 and 2 is further introduced below in conjunction with specific example, see the following description for details:
[0076] (1) Data preparation:
[0077] (a) Divide the dataset
[0078] The samples were divided into different sets according to the class, and the data sources were 96 cases of undisclosed breast tumor MRI, including: 27 cases of malignant tumor samples and 69 cases of benign tumor samples.
[0079] Randomly select 5 cases in the malignant tumor samples (positive samples), and randomly select 10 cases in the benign tumor samples (negative samples) as the verification set; the rest of the data are used as the training set.
[0080] (b) Data preprocessing
[0081] Each 3D MRI sample was cropped to get a 2D image of the tumor area, and all 2D images were normalized to a uniform size of 224×224.
[0082] Finally, 1847 training set data (including 387 positive samples and 1460 negative samples) and 365 verification set data (inclu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com