A kind of novel chimeric dna polymerase and preparation method thereof
A polymerase and exonuclease technology, applied in the field of DNA polymerase, can solve the problems of low continuous synthesis ability, slow amplification and extension speed of Pfu enzyme, and low error probability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1: Design of chimeric DNA polymerase
[0041] Pfu and KOD have very different phenotypic characteristics, in particular, in terms of elongation rate, processivity, and error rate, Pfu has the lowest error rate among all thermostable polymerases, with an error rate of about 2.0 x10 -6, KOD is a DNA polymerase with high amplification ability, the amplification speed is 2 times that of Taq enzyme, 6 times that of Pfu enzyme, and the amplification yield is higher (~300nts) (takagi et al.1997).
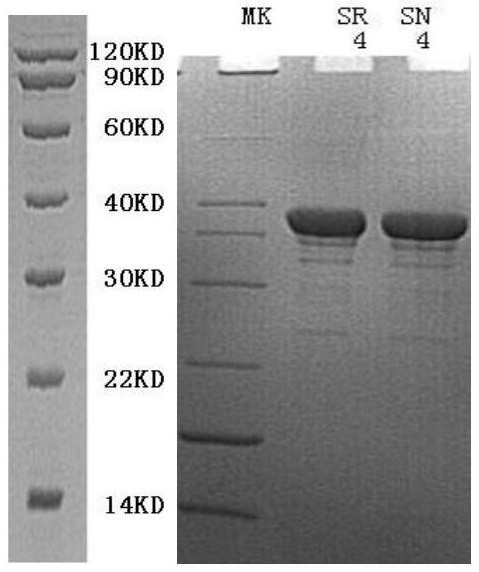
[0042] Therefore, the chimeric DNA polymerase of this example is a chimeric combination of these two enzymes, which exhibits a lower error rate (0.93×I0-6) than Pfu, and has processivity and elongation comparable to KOD rate (about 300nt / s and 106-138nt / s, respectively). The nucleotide sequence of the chimeric DNA polymerase provided by the present invention is SEQ ID No.3, specifically the N-terminal domain nucleotide sequence (1-390&981-1104) of the KOD enzyme, the pa...
Embodiment 2
[0043] Example 2: Purification of Chimeric DNA Polymerase
[0044] Inoculate the obtained expression strain 1:100 into LB medium, culture at 37°C, 250rmp shaking until OD600=2.5, add 0.1mM IPTG to induce expression, in order to increase the ratio of protein soluble expression, reduce the culture temperature to 16 ℃, low temperature induced expression overnight (16h), 5k centrifugation harvested bacteria.
[0045] 1. Bacteria destruction: the total amount of bacteria is mixed with the bacteria-breaking buffer (50mM Tis-Hcl, pH 8.0) at 1:10, that is, 1g of bacteria is added to 10ml of buffer, and the bacteria are broken by ultrasonic. The supernatant was collected by centrifugation at 8000rpm for 30min.
[0046] 2. Mix the thermal denaturation buffer (25mM Tris, 1mM EDTA, 0.5% Tween20, pH8.0,) into the ultrasonically sterilized solution at a ratio of 1:1, mix well and put it in a water bath at 75°C for 30 minutes to denature. The supernatant was collected by centrifugation at ...
Embodiment 3
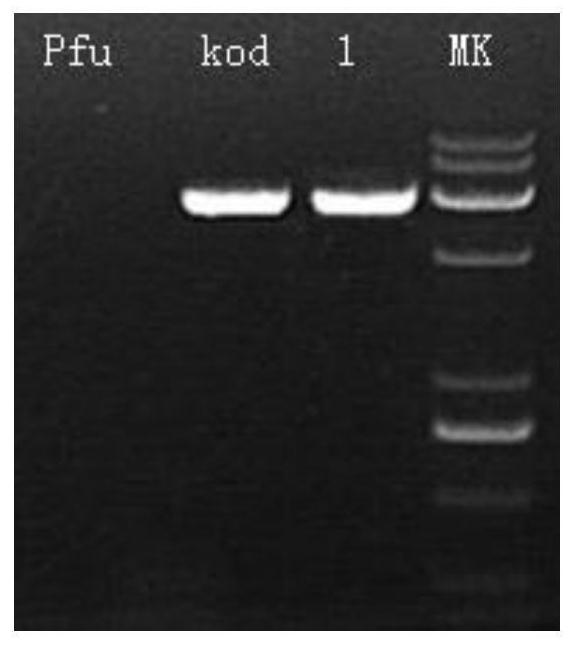
[0049] Example 3: Detection of extension rate of chimeric DNA polymerase
[0050] The chimeric high-fidelity DNA polymerase, Pfu DNA polymerase and Kod DNA polymerase prepared in Example 1 of the present invention were amplified using λDNA as a template, and the amplified fragment was 3000 bp. The result is as image 3 Shown: Lnae1 is a chimeric DNA polymerase.
[0051] The reaction system and procedure are as follows:
[0052]
[0053]
[0054] Example 3: Fidelity Detection of Chimeric DNA Polymerase
[0055] 1) Amplify the LacIZα gene, the primer sequence is as follows:
[0056] F: GTTTTCCCAGTCACGAC
[0057] R: GGTATCTTTATAGTCCTGTCG
[0058] 2) Endonuclease digestion to obtain a linearized vector:
[0059] 3) Ligate the obtained gene to the vector, transform the ligated product into the host strain DH5a for expression, culture it on a medium containing x-gal and IPTG, count the number of blue and white spots, and calculate the fidelity.
[0060] Calculated accor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com