DNA extracting agent, DNA extracting method and DNA extracting kit
An extraction method and kit technology, applied in DNA preparation, recombinant DNA technology, etc., can solve the problems of low DNA yield, affecting PCR, high-throughput sequencing, low DNA purity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment
[0041] The following will be described in detail in conjunction with specific embodiments. The following examples, unless otherwise specified, do not include other components except unavoidable impurities. The drugs and instruments used in the examples are all routine choices in the art unless otherwise specified. The experimental methods for which specific conditions are not indicated in the examples are implemented according to conventional conditions, such as the conditions described in literature, books or the method recommended by the manufacturer.
Embodiment 1
[0044] Weigh three stool samples from the same person, each 200 mg, and perform the following operations for each stool sample:
[0045] (1) Pretreatment: Mix the feces sample with 5 times the volume of normal saline, vortex to mix, centrifuge at 2000rpm / min at 4°C for 2min, take the supernatant and discard the precipitate, then 10000rpm / min at 4°C Centrifuge at a high speed for 1 min, discard the supernatant, and keep the centrifuged pellet. Then add 1mL PBS (pH 7.4) to the precipitate, vortex and mix well, centrifuge at 2000rpm / min for 2min at 4°C, take the centrifuged supernatant and discard the precipitate; then put the centrifuged supernatant at 4 Centrifuge at 10,000 rpm / min for 1 min at ℃, discard the supernatant, and keep the precipitate. At this time, the precipitate is the purified microbial cell, which is the sample to be lysed.
[0046] (2) Lysis solution to lyse the sample: add 800 μL of lysis solution to the sample to be lysed obtained in step (1), vortex to mix...
Embodiment 2
[0052] The steps and methods for extracting DNA in the feces in Example 2 are roughly the same as in Example 1, except that the lysate used in Example 2 is composed of guanidine isothiocyanate with a final concentration of 1M and Tris- It is composed of HCl, N-lauroyl sarcosine with a final mass volume percentage of 5%, and a phosphate buffer solution with a final concentration of 0.05M pH8.0. TENP buffer was composed of 100 mM Tris pH 8.0, 50 mM EDTA pH 8.0, 500 mM NaCl and 6% PVPP in final mass volume. The first wash consisted of phosphate at a final concentration of 0.05M pH 8.0 and potassium acetate at a final concentration of 1.5M.
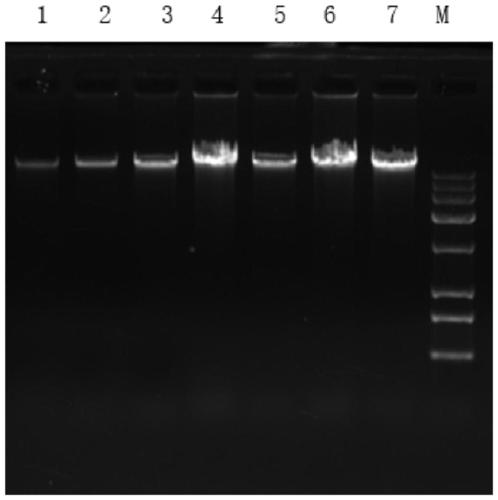
[0053] The concentration of DNA in three groups of stool samples of embodiment 2 is shown in table 1 respectively, and electrophoresis figure is as shown in Table 1. figure 1 shown. In Table 1, numbers 5-7 are the extraction results of the three stool samples in Example 2. figure 1 The number 1 in the middle corresponds to the E.coli DNA s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com