Application of circ_ADARB1 in preparing nasopharyngeal carcinoma therapeutic preparation and therapeutic preparation
A technology for nasopharyngeal carcinoma and preparations, which is applied in the field of tumor molecular biology and can solve the problems of unclear molecular mechanism and insignificant curative effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] Example 1: Expression of circ_ADARB1 in nasopharyngeal carcinoma tissues and cells
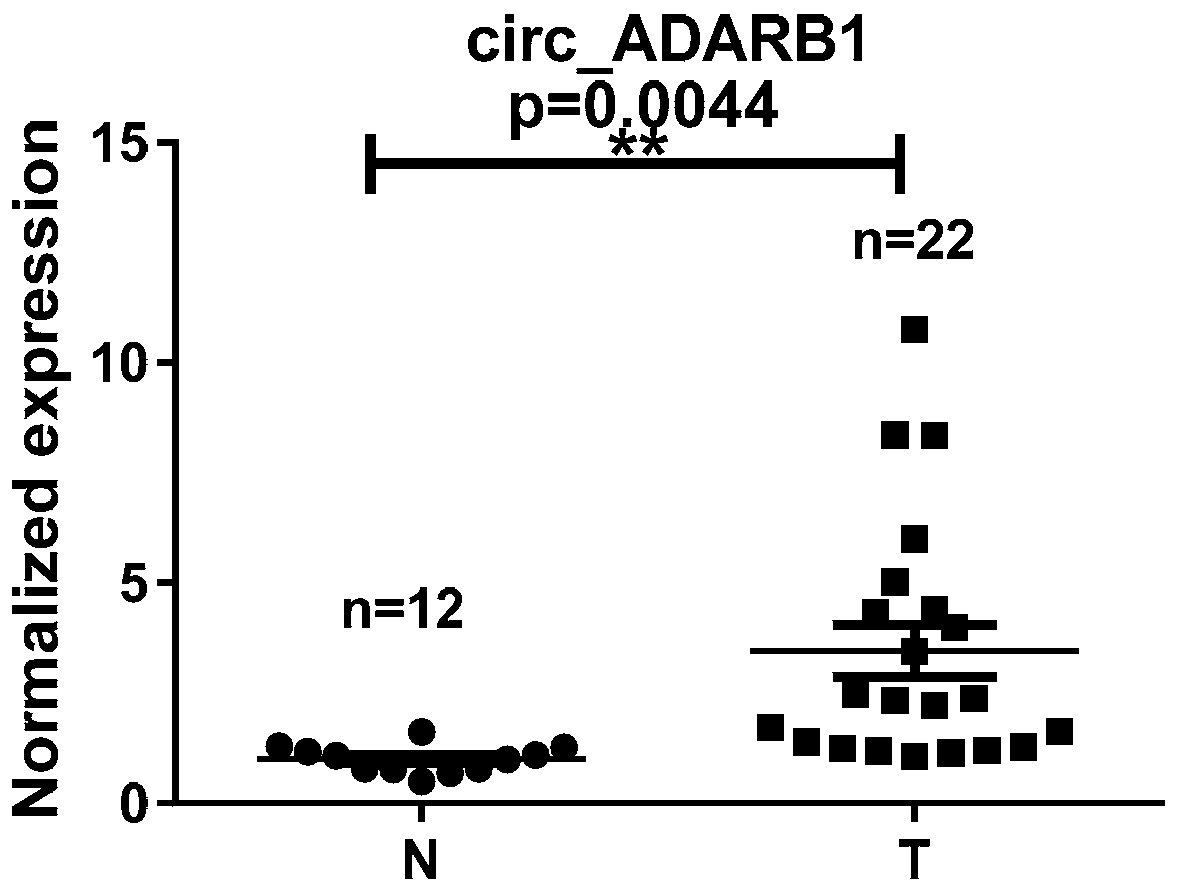
[0092] 1. According to the standard sample collection plan, we collected 34 tissue samples from patients with nasopharyngeal carcinoma from Hunan Cancer Hospital. All the cases were newly diagnosed patients in the Department of Head and Neck Surgery of Hunan Cancer Hospital (time interval: January 2016 to November 2016). 22 cases of nasopharyngeal carcinoma and 12 cases of nasopharyngeal inflammation were diagnosed by the pathology department (neoplastic diseases, no active infectious diseases, severe immune diseases and other major diseases have been excluded).
[0093] Complete personal information and clinical data were recorded during the collection process, including name, gender, age, outpatient number, hospitalization number, pathological type, case stage, and EBV infection, etc. See the screenshot of the Excel spreadsheet for details. All samples are collected with the permissi...
Embodiment 2
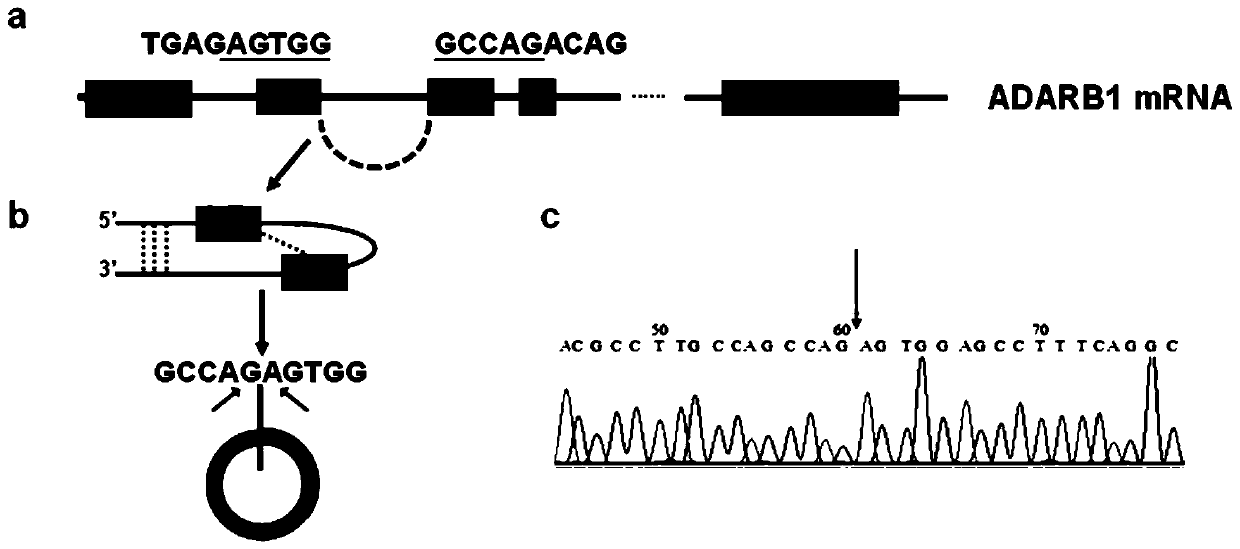
[0129] Example 2: Sanger sequencing proves that circular RNA is formed
[0130] In order to prove that circ_ADARB1 forms a circular RNA rather than a linear one, the figure 1 The qRT-PCR products were recovered and sent to the company for sanger sequencing (Qingke Company). Compare the sequences returned by the company with DNASTAR software, and use the chromas software to view the peak diagrams to judge the quality of the sequencing. figure 2 The results in the middle show that a. circ_ADARB1 is formed by end-to-end splicing of exons 2 and 3 of ARHGAP12, E indicates exon (exon), and the underlined sequence indicates the linker sequence at the end; b. Schematic diagram of circRNA formation; c. Peak of sequencing results In the figure, the black arrow indicates the end-to-end connection from here.
Embodiment 3
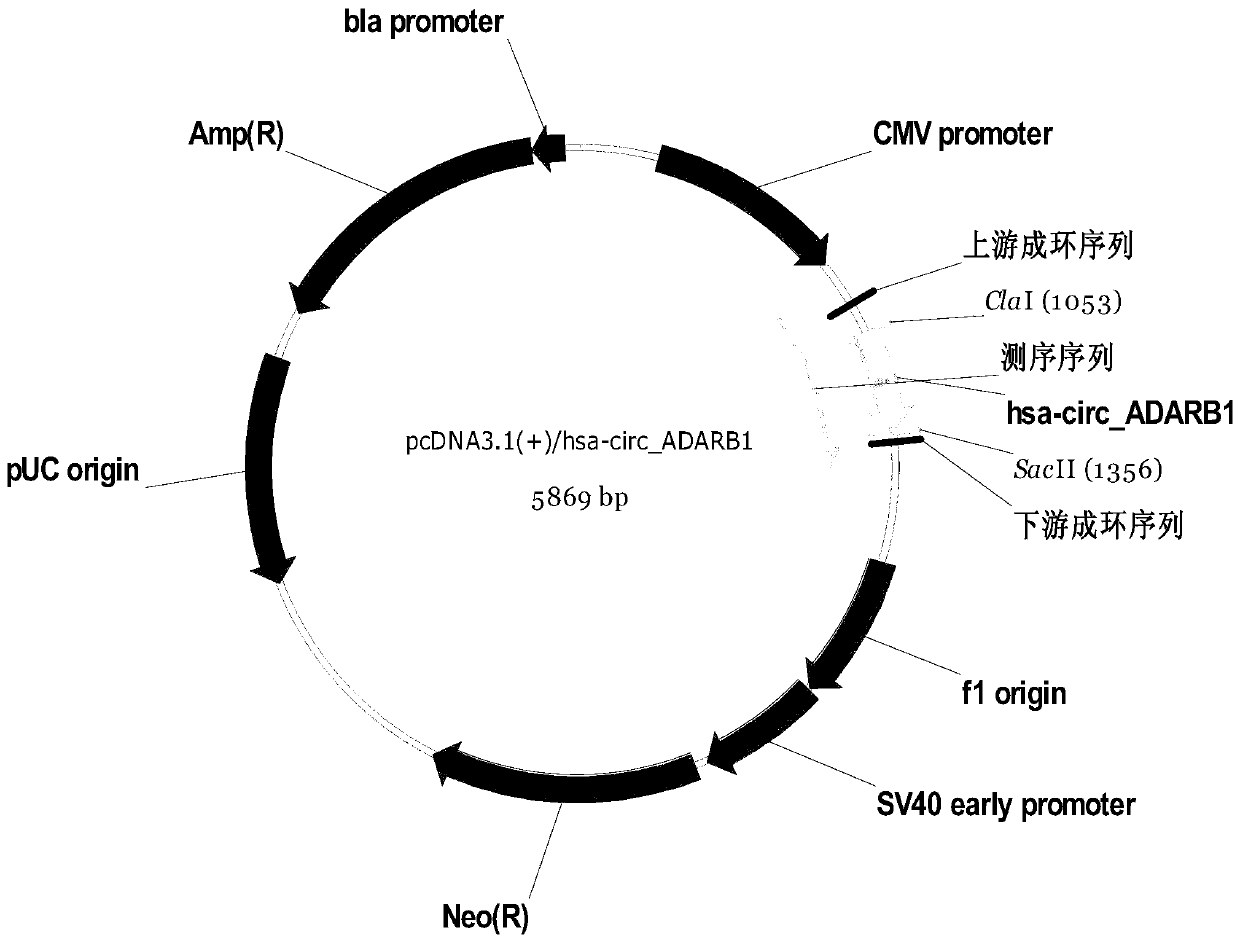
[0131] Example 3: Detection of the overexpression effect of circ_ADARB1 in nasopharyngeal carcinoma cell lines
[0132]First, we select the restriction site, and put the full-length sequence of circ_ADARB1 into the NEB cutter 2.0 online website for analysis. It shows that the ClaI and SacII restriction sites are sites that do not exist in the full-length sequence of circ_ADARB1, and at the same time in the pcDNA3.1 plasmid vector A single DNA restriction enzyme. Construct the overexpression body accordingly; image 3 The overexpression vector map drawn for.
[0133] In order to test the circling efficiency of circ_ADARB1, we first expressed the constructed pcDNA3.1 / circ_ADARB1 eukaryotic overexpression vector in nasopharyngeal carcinoma cells. The third and fourth generation nasopharyngeal carcinoma cells CNE2 and HNE2 in good growth condition were seeded into 12-well plates, and when the cell confluence reached 60%-80%, the endotoxin-free plasmid pcDNA3. 1 Empty vector and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com