Method for identifying Hucho taimen and Brachymystax lenok
A technology for salmonids and salmonids, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as mixed germplasm, achieve simple operation, improve work efficiency, and save money The effect of cost and time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0012] Specific implementation mode one: the method for identifying the genetic sex of salmon in this implementation mode:
[0013] 1. Extract the genomic DNA of the sample to be identified;
[0014] 2. Perform PCR amplification using the genomic DNA in step 1 as a template. In the PCR amplification system, the upstream primer of the discrimination primer pair is 5'-ATCAATCATGGCTGGAGGTG-3', and the downstream primer of the discrimination primer pair is 5'-CACCCACACACGTACCAGTC-3';
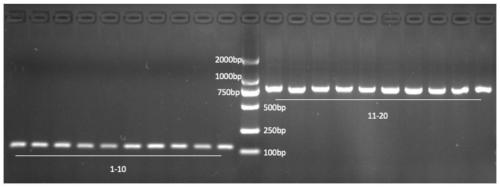
[0015] 3. Gel electrophoresis of the PCR product. If the amplified band size is 133bp, it is called salmon, and if the amplified band size is 814bp, it is called salmon.
specific Embodiment approach 2
[0016] Embodiment 2: The difference between this embodiment and Embodiment 1 is that the PCR amplification reaction system in step 2 is 20 μl: 2×Taq PCR mix 10 μl, the genomic DNA of the sample to be identified is 2 μl, the concentration of the discrimination primer is 10 μM, and the discrimination primer 1 μl each of the upstream and downstream primers of the pair, and the rest were supplemented with ultrapure water; the PCR reaction was: pre-denaturation at 95°C for 3 minutes, followed by 32 cycles of denaturation at 95°C for 30 sec, annealing at 66°C for 30 sec, and extension at 72°C for 30 sec; finally, extension at 72°C 5min. Others are the same as the first embodiment.
[0017] Under the PCR amplification reaction conditions of this embodiment, the target sequence is completely denatured without affecting the activity of the Taq enzyme, the primers are completely extended, an effective amplification amount can be achieved, and the non-specific amplification is small.
specific Embodiment approach 3
[0018] Specific embodiment three: the difference between this embodiment and specific embodiment one or two is:
[0019] Step 1: Take 5-10 scales or 0.4cm 2 The fin rays were lysed with 40 μl lysate, and then digested with a PCR instrument. The digestion process was 55°C for 50 minutes and 98°C for 10 minutes; after that, mix well, and then centrifuge at 1000-2000rpm for 1-2 minutes, and take the supernatant to obtain the sample Genomic DNA. Others are the same as the first or second embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com