Binding assay
A technology of binding activity and LAG-3, applied in the field of probes and kits, can solve the problems that cannot be used to determine the specific binding of IMP321 to immobilized cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Evaluation of the use of a fluorescence-activated cell sorting (FACS) assay for determining the binding of IMP321 to Raji cells
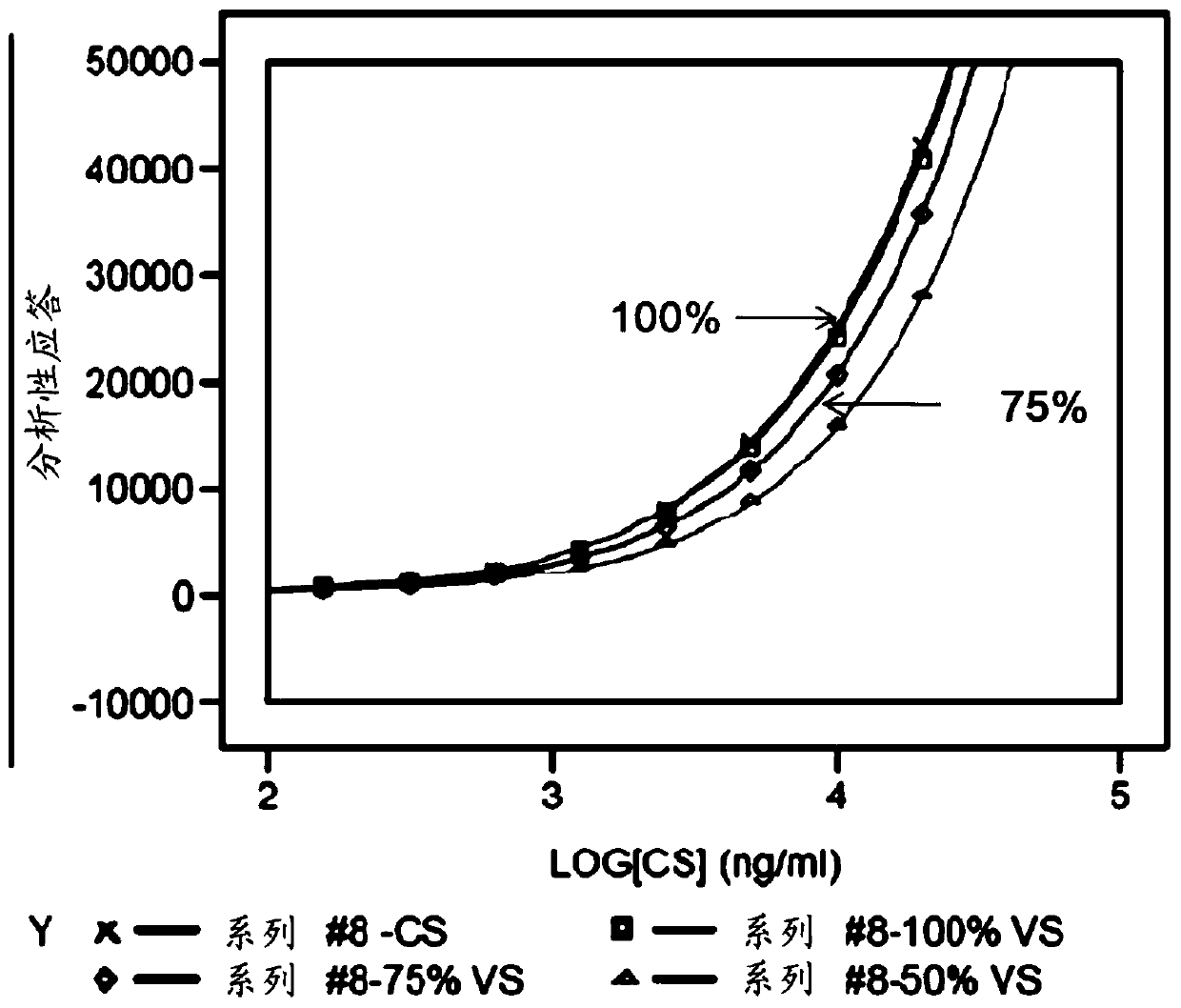
[0097] FACS assays were performed to determine the binding of IMP321 to Raji cells. IMP321 samples with 100%, 75% and 50% MHC class II binding activity were tested. A sample with 100% activity is a reference sample with known MHC class II binding activity at a predetermined concentration. Samples with 75% and 50% activity were prepared by diluting the reference sample.
[0098] The obtained binding curves are shown in figure 2 middle. They show that no upper plateau is reached, so there is no parallelism between the binding curves of the reference sample with 100% activity and the other samples. This prevents calculation of relative potencies of different samples.
Embodiment 2
[0100] Evaluation of the use of the Meso Scale Discovery (MSD) assay for determining the binding of IMP321 to Raji cells
[0101] This example describes the evaluation of the Meso Scale Discovery (MSD) assay used to determine the binding of IMP321 to Raji cells.
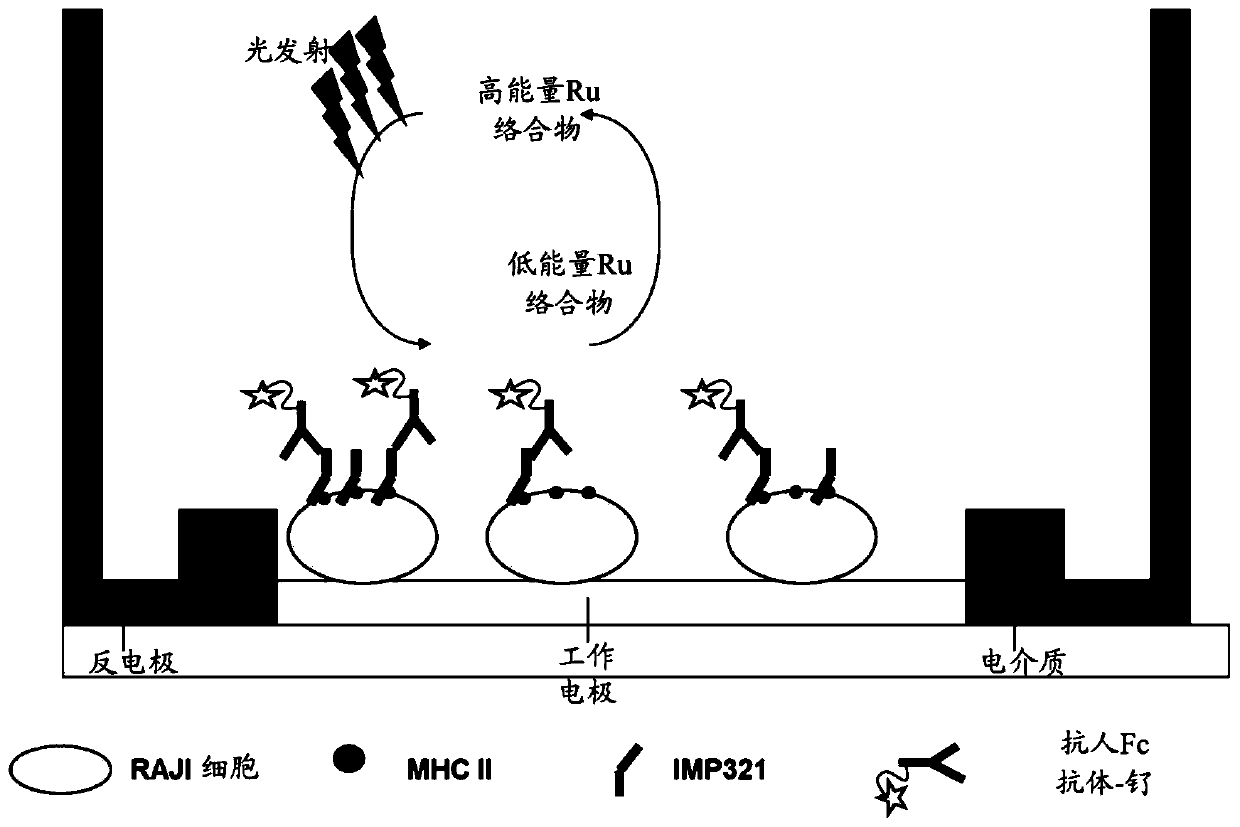
[0102] The Meso Scale Discovery platform (MSD-ECL) uses electrochemiluminescent labels conjugated to detection antibodies. When electrically stimulated in the appropriate chemical environment, these tags generate light, which can then be used to measure key proteins and molecules.
[0103] Power is applied to the plate electrodes through the Meso Scale Discovery platform (MSD-ECL), causing the markers to emit light. The light intensity is then measured to quantify the analyte in the sample.
[0104] The detection process starts at electrodes located in the bottom of a Meso Scale Discovery (MSD-ECL) microplate, and only labels near the electrodes are excited and detected. The system employs a buffer solution with...
Embodiment 3
[0111] Evaluation of non-specific binding of IMP321 to ELISA plates
[0112] This example describes the evaluation of non-specific binding of IMP321 and Rituxan to plates for enzyme-linked immunosorbent assay (ELISA) using different blocking reagents.
[0113] Briefly, microplates were blocked with blocking reagent for 2 hours at 25°C. Samples and rituxan controls were diluted to 2 μg / ml with dilution buffer and then further diluted by two-fold serial dilution. Before and after addition of diluted samples and incubation, the microplates were washed and drained well. After incubation with secondary antibodies, the signal was measured by spectrometry using a SpectraMax M2 (450-650 nm).
[0114]
[0115] The test results are in Figure 5 shown in . Figure 5 (a) shows the results of ELISA using increasing concentrations of IMP321 and ELISA plates blocked with 5% BSA or 10% FBS. Figure 5 (b) shows the results of ELISA using increasing concentrations of IMP321 or Rituxan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com