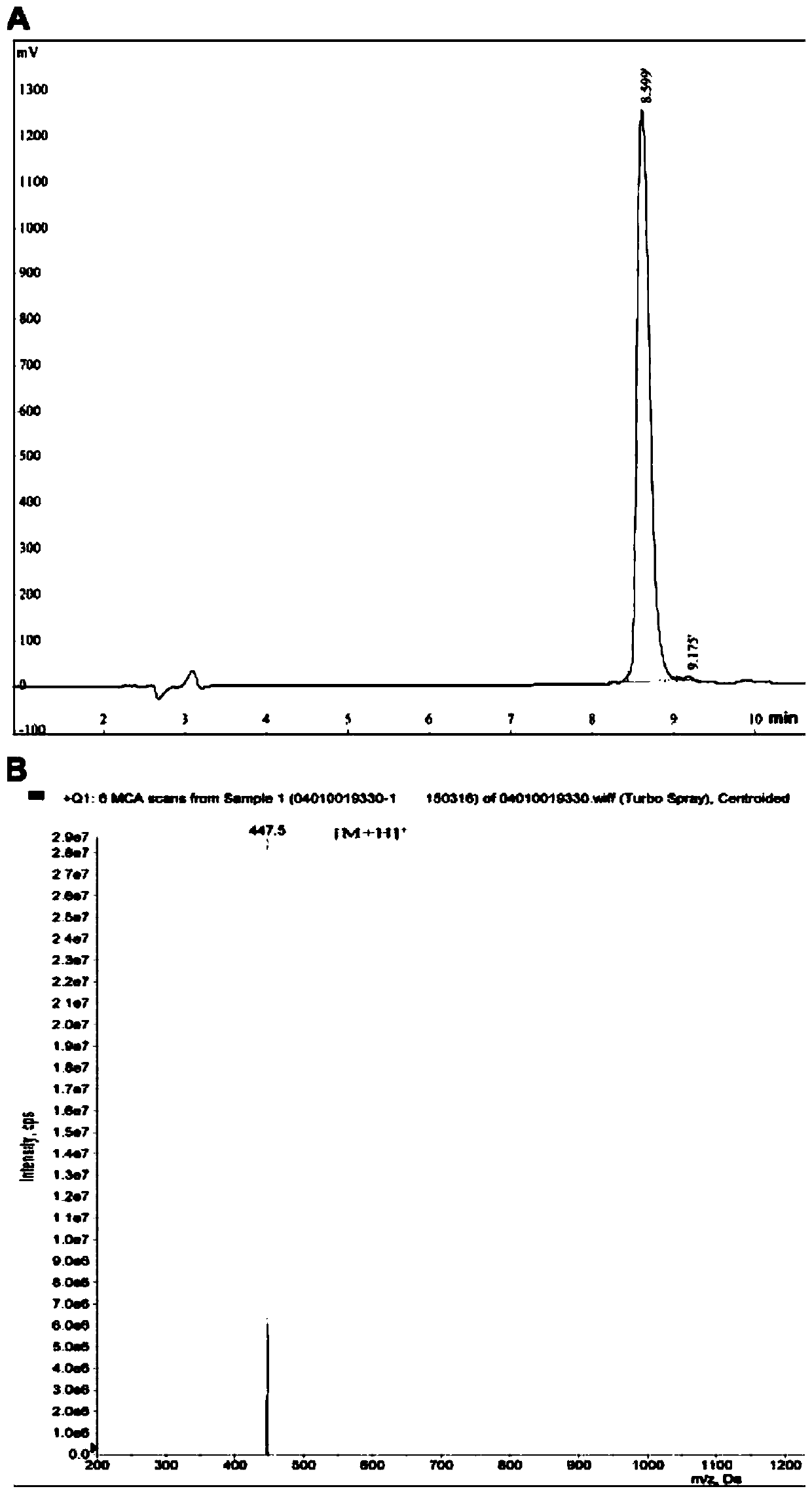
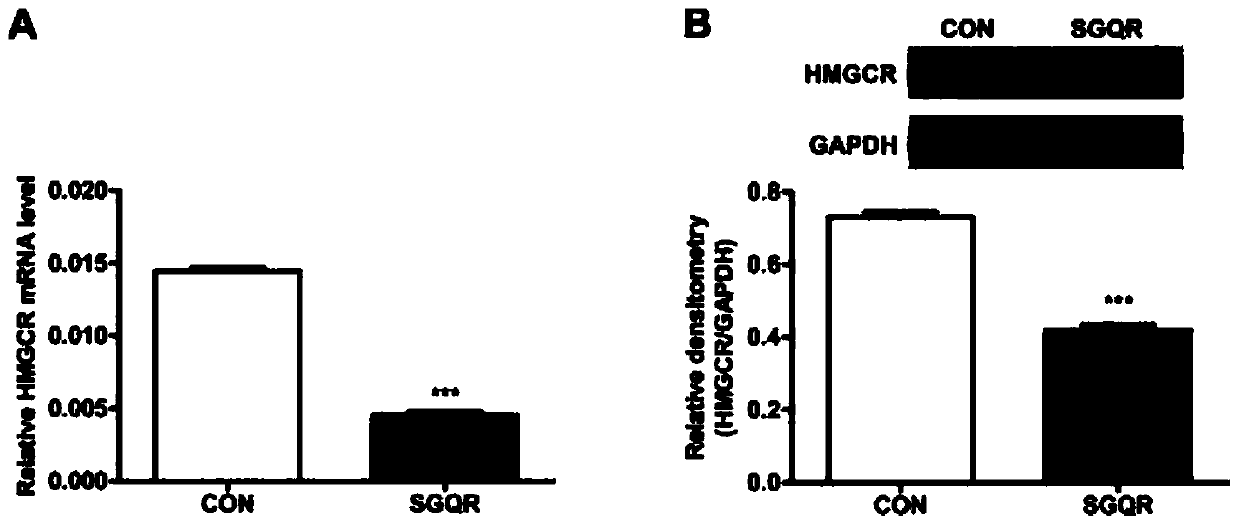
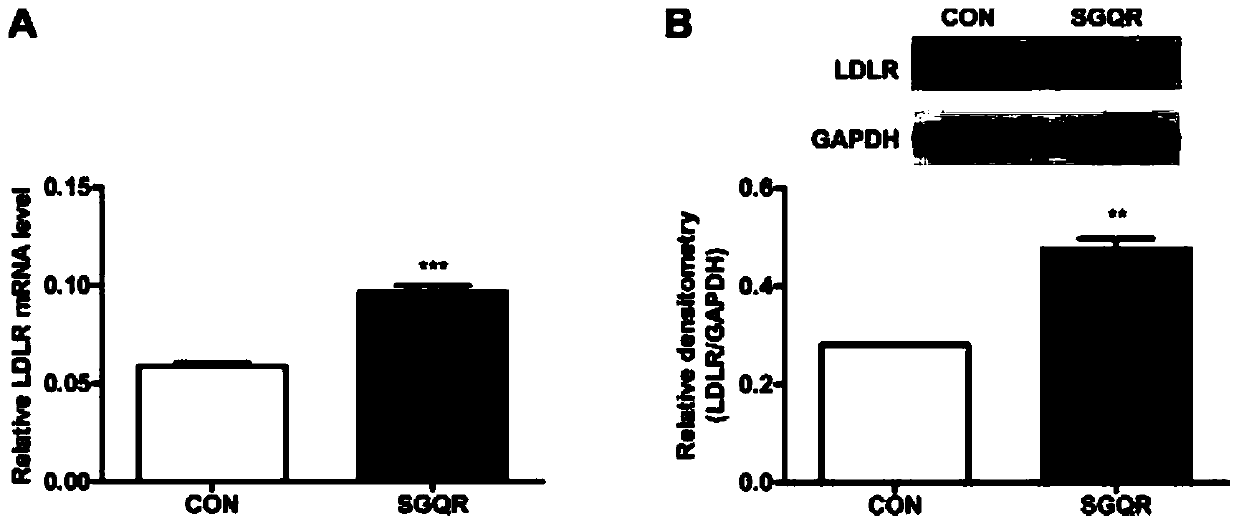
Silkworm pupa protein source tetrapeptide SGQR and application thereof
A technology of silkworm chrysalis protein and small molecule peptides, which is applied in the field of tetrapeptide Ser-Gly-Gln-Arg, can solve the problem that the mechanism of action is not clear, and achieve the effect of inhibiting the expression level of SQS gene and protein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] (1) Inoculate HepG2 cells into a 6-well plate and culture for 24 hours. After discarding the old medium, add culture medium with a final concentration of 0 (blank control group) and 500ng / mL SGQR (experimental group) to each well. cultured for 12 hours respectively. The above cell culture conditions were 37°C, CO 2 Content 5%, relative humidity 95%. The medium formula is: high glucose DMEM medium containing 10% FBS, 1% penicillin / streptomycin.
[0050] (2) After the end of the culture, discard the culture medium in each well in (1), wash the cells three times with ice-cold PBS, then add 1.0ml Trizol to each well to lyse the cells, then transfer to a centrifuge tube, add 200 μl chloroform; vigorously Shake for 15Sec (avoid shaking with a shaker), let stand at room temperature for 2min, then centrifuge at 12000g for 10min at 4°C; use a pipette to draw the upper aqueous phase into another clean centrifuge tube, add 600μL of isopropanol and mix upside down, 12000g 4 Cent...
Embodiment 2
[0056] (1) Inoculate HepG2 cells in a 6-well plate and culture for 24 hours. After discarding the old medium, add culture medium with a final concentration of 0 (blank control group) and 500ng / mL SGQR (experimental group) to each well. cultured for 24 hours respectively. The above cell culture conditions are all at 37°C, with a CO2 content of 5%, and a relative humidity of 95%. The medium formula is: high glucose DMEM medium containing 10% FBS, 1% penicillin / streptomycin.
[0057] (2) Remove the culture medium from the cells in each well in (1), wash 3 times with iced PBS, add 100 μL of lysate containing protease and phosphatase inhibitors and lyse on ice for 30 minutes, scrape the cells into 1.5mL EP In the tube, heat above 95°C for 10 minutes, then centrifuge at 12,000 rpm for 10 minutes at 4°C, take the supernatant, quantify the protein, and store it in a -80°C refrigerator.
[0058] (3) Load the protein lysate in (2), first electrophoresis the stacking gel at 80V for 20m...
Embodiment 3
[0064] (1) Use the Docking module in the MOE software to carry out the molecular docking of the active peptide and HMGCR. The HMGCR structure was obtained from the crystal structure in the PDB biomacromolecular structure database (PDB code: 1HW8).
[0065] (2) Use the Docking module in the MOE software to carry out the molecular docking of the active peptide and LDLR. The LDLR structure was obtained from the crystal structure in the PDB biomacromolecular structure database (PDB code: 2MG9).
[0066] (3) Use the Docking module in the MOE software to carry out the molecular docking of the active peptide and SQS. The SQS structure was obtained from the crystal structure in the PDB biomacromolecular structure database (PDB code: 3WC9).
[0067] The molecular docking technique was used to analyze the interaction mechanism of SGQR with HMGCR, LDLR and SQS. SGQR and HMGCR can combine with four hydrogen bonds, which are the three amino acids Glu A730, Asn A734 and Glu C782 in the a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com