Methods of using archaeal serine protease
A serine protease and amino acid technology, applied in the field of thermophilic protease, can solve problems such as stability or harmful activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0427] Unless defined otherwise herein, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this disclosure belongs. Singleton et al., DICTIONARY OFMICROBIOLOGY AND MOLECULAR BIOLOGY, 2nd ed., John Wiley and Sons, New York (1994), and Hale and Marham, THE HARPER COLLINSDICTIONARY OF BIOLOGY [ HarperCollins Biological Dictionary], Harper Perennial, New York (1991) provides the skilled artisan with a general dictionary of many of the terms used in this disclosure.
[0428] The disclosure is further defined in the examples below. It should be understood that these Examples, while illustrating certain embodiments, are given by way of illustration only. From the above discussion and these Examples, one skilled in the art can ascertain the essential characteristics of this disclosure, and without departing from the spirit and scope of this disclosure, can make various changes and modifications to a...
example 1
[0430] Analysis of the TnaPro1 sequence
[0431] The nucleotide sequence of the full-length TnaPro1 gene identified in the NCBI database is provided in SEQ ID NO: 1 (Thermophilus nautilus, NCBI Reference Sequence: NZ_CP007264.1, 1327825-1329105, complement). The corresponding full-length protein sequence encoded by the TnaPro1 gene is shown in SEQ ID NO: 2 (GenBank accession number: AHL23118.1).
[0432] The propeptide and mature regions of the TnaPro1 protein were predicted based on the protein sequence alignment with the homologous enzyme Thermococcus kodakaraensis KOD1 TK-subtilisin protein (Pulido et al. (2006) Applied and Environmental Microbiology, 72( 6): 4154-4162). Using SignalP v4.0 (Nordahl Petersen et al. (2011) Nature Methods [Nature Methods] 8:785-786) software, the TnaPro1 signal peptide is predicted to have the sequence shown in SEQ ID NO: 10, and the zymogen is predicted to have the sequence shown in SEQ ID NO: 10 The sequence shown in ID NO:8. The N-termin...
example 2
[0435] Cloning of TnaPro1
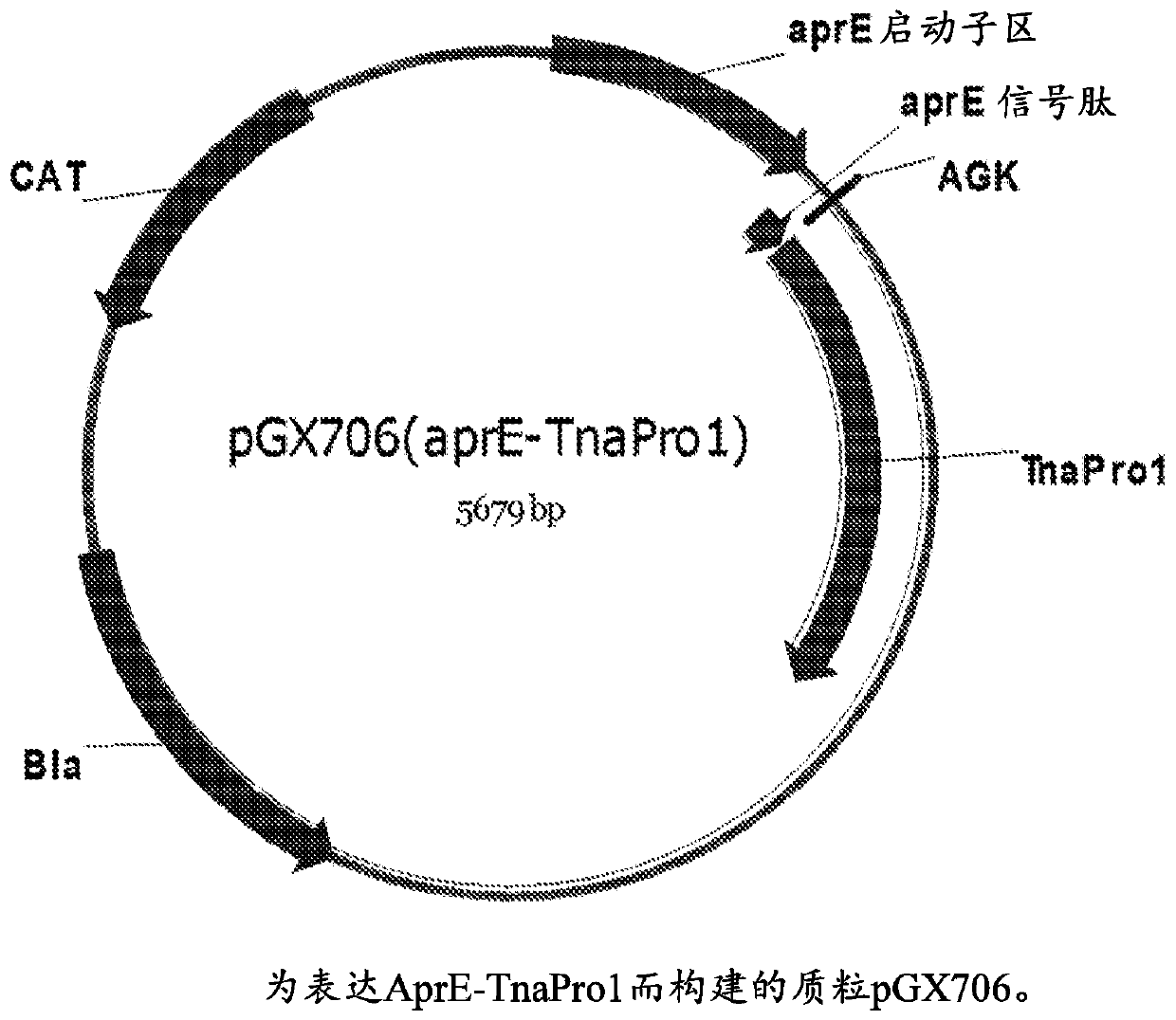
[0436] The DNA sequence encoding the proenzyme of TnaPro1 was synthesized and inserted into the Bacillus subtilis expression vector p2JM103BBI (Vogtentanz, Protein Expr Purif [protein expression and purification], 55:40-52, 2007) to obtain the name pGX706 (aprE- TnaPro1) plasmid ( figure 1 ).
[0437] Ligation of the gene encoding the TnaPro1 protein into the digested vector resulted in the addition of three codons (encoding Ala-Gly-Lys ). This gene has an alternative start codon (GTG). Such as figure 1 As shown, pGX706(aprE-TnaPro1) contains an AprE promoter, an AprE signal sequence for directing Bacillus subtilis to secrete the target protein, and a synthetic nucleotide sequence encoding the predicted TnaPro1 zymogen. SEQ ID NO: 4 shows the gene sequence of the aprE-TnaPro1 construct, including three additional codons introduced during ligation of the fusion sequence. SEQ ID NO:5 shows the amino acid sequence of the AprE-TnaPro1 fusion pro...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| water activity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com