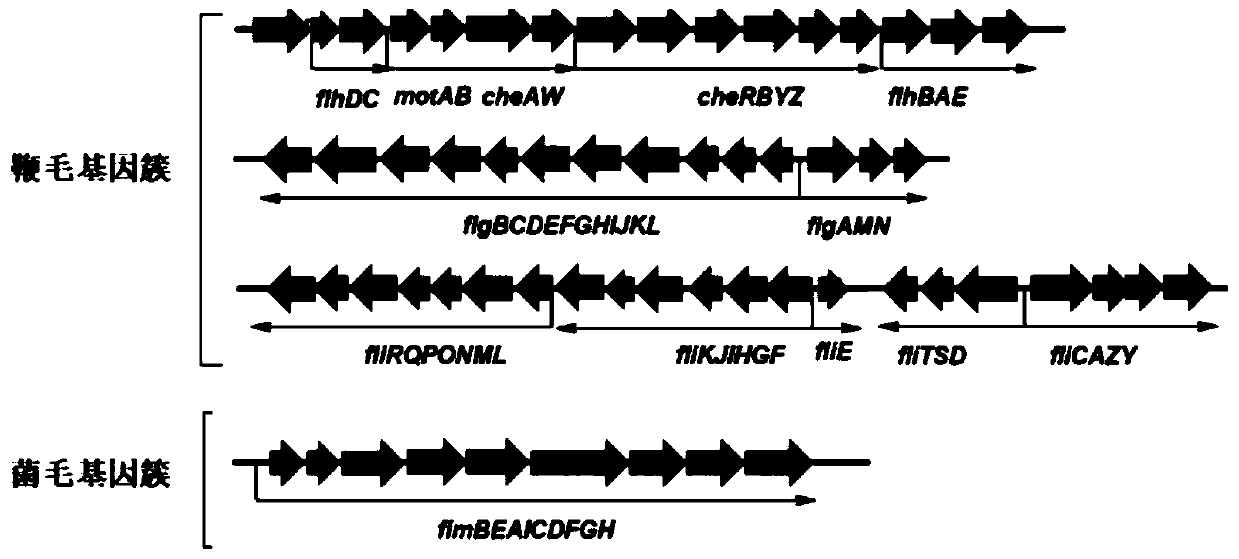
Method for efficiently synthesizing PHB by knocking out flagellum and pilus genes
A flagella and gene technology, applied in the field of efficient synthesis of PHB, can solve the problems such as the inability of PHB to meet the needs of industrial production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Construction of embodiment 1 knockout plasmid
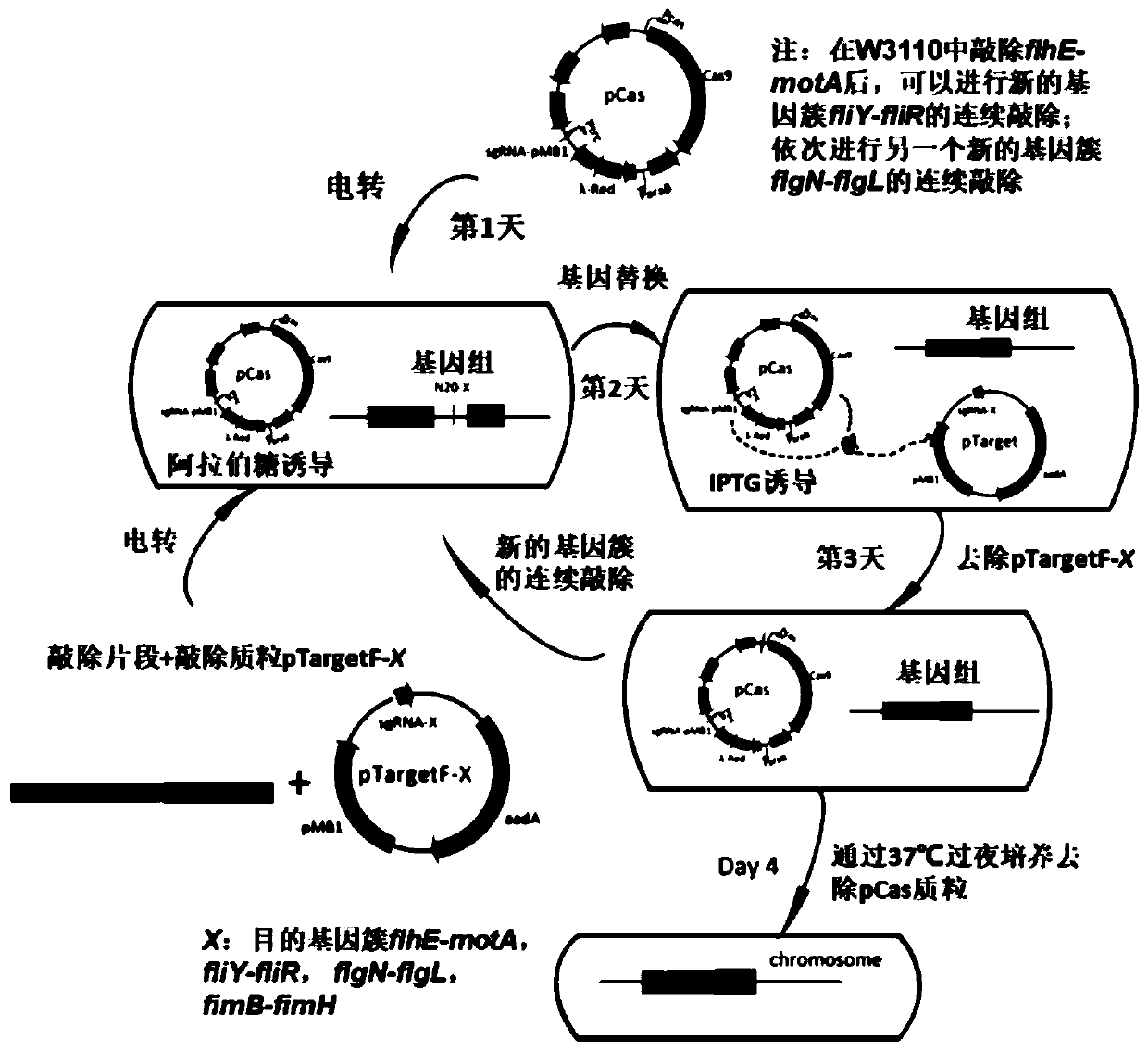
[0051] Using the CRISPR / Cas9 knockout system to knock out flagella and pilus synthesis and transport gene clusters, four knockout plasmids need to be constructed: pTargetF-flhD-motA, pTargetF-fliY-fliR, pTargetF-flgN-flgL, pTargetF-fimB-fimH , the construction process of the plasmid is as follows:
[0052] (1) Through the analysis of the website http: / / crispr.mit.edu / , select a 20nt N that is complementary to the target sequence of the target gene 20 sequence, specific N 20 The sequences are the underlined sequences of the primers pTargetF-flhE-motA-F, pTargetF-fliY-fliR-F, pTargetF-flgN-flgL-F, and pTargetF-fimB-fimH-F, and these sequences are modified into the plasmid pTargetF forward primer At the 5' end, forward primers pTargetF-flhD-motA-F, pTargetF-fliY-fliR-F, pTargetF-flgN-flgL-F, and pTargetF-fimB-fimH-F were respectively obtained. Using the plasmid pTargetF as a template, the forward primers pTargetF-flhD-motA...
Embodiment 2
[0059] The construction of embodiment 2 genetically engineered bacteria WJW010 and WJW011
[0060] The CRISPR / Cas9 knockout system was used to knock out the flagella synthesis and transport gene cluster of wild-type Escherichia coli W3110 to obtain engineering strain WJW010; the pili gene cluster was knocked out to get WJW011. The specific knockout process is as follows:
[0061] (1) Preparation of Escherichia coli electroporation knockout competent
[0062] The plasmid pCas is transformed into Escherichia coli W3110 to obtain recombinant Escherichia coli W3110 / pCas containing the pCas plasmid, and the Escherichia coli W3110 / pCas is activated on the LB solid plate adding 30mg / L Kanamycin (Kan), and inoculated into LB +Kan test tube cultured overnight to obtain seed liquid; transfer the seed liquid to 25mL LB+Kan medium at 1%, and cultivate to OD at 30°C and 200rpm 600 =0.2, add 500 μL L-arabinose solution to induce, continue to culture to OD 600 =0.5, ice bath for 30min; 4°...
Embodiment 3
[0075] Example 3 Growth status of flagella-reduced strain WJW010 and pilus-reduced strain WJW011
[0076] The streamlined strains WJW010 and WJW011 were 20, 6440bp and 8753bp streamlined respectively compared with the wild type W3110, and the streamlined genome proportions were about 1.13% and 0.19% respectively. W3110, WJW010 and WJW011 were cultured, and their growth curves in LB medium were determined.
[0077] The results showed that the growth curve as Figure 5 As shown, all three entered the stable phase at 10 h; in the early logarithmic period, the growth of the strains WJW010 and WJW011 was slightly slower than that of the wild type; It was basically the same as the wild type W3110; after the stable period, the OD of the streamlined strains WJW010 and WJW011 600 Taller, highest OD of the flagellar-reduced strain WJW010 600 Compared with W3110, it increased by 12.4%, and finally increased by 10.7%; the pili reduced strain WJW011 had the highest OD 600 A 6.6% improv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com