Method for analyzing peptide fragment impurities in high-purity polypeptide based on data mining
A technology of data mining and impurity analysis, applied in the direction of analyzing materials, material separation, measuring devices, etc., can solve the problems of insufficient flexibility, lack of solutions, and complex MS/MS data spectrum, so as to increase the number of identifications and increase coverage. rate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
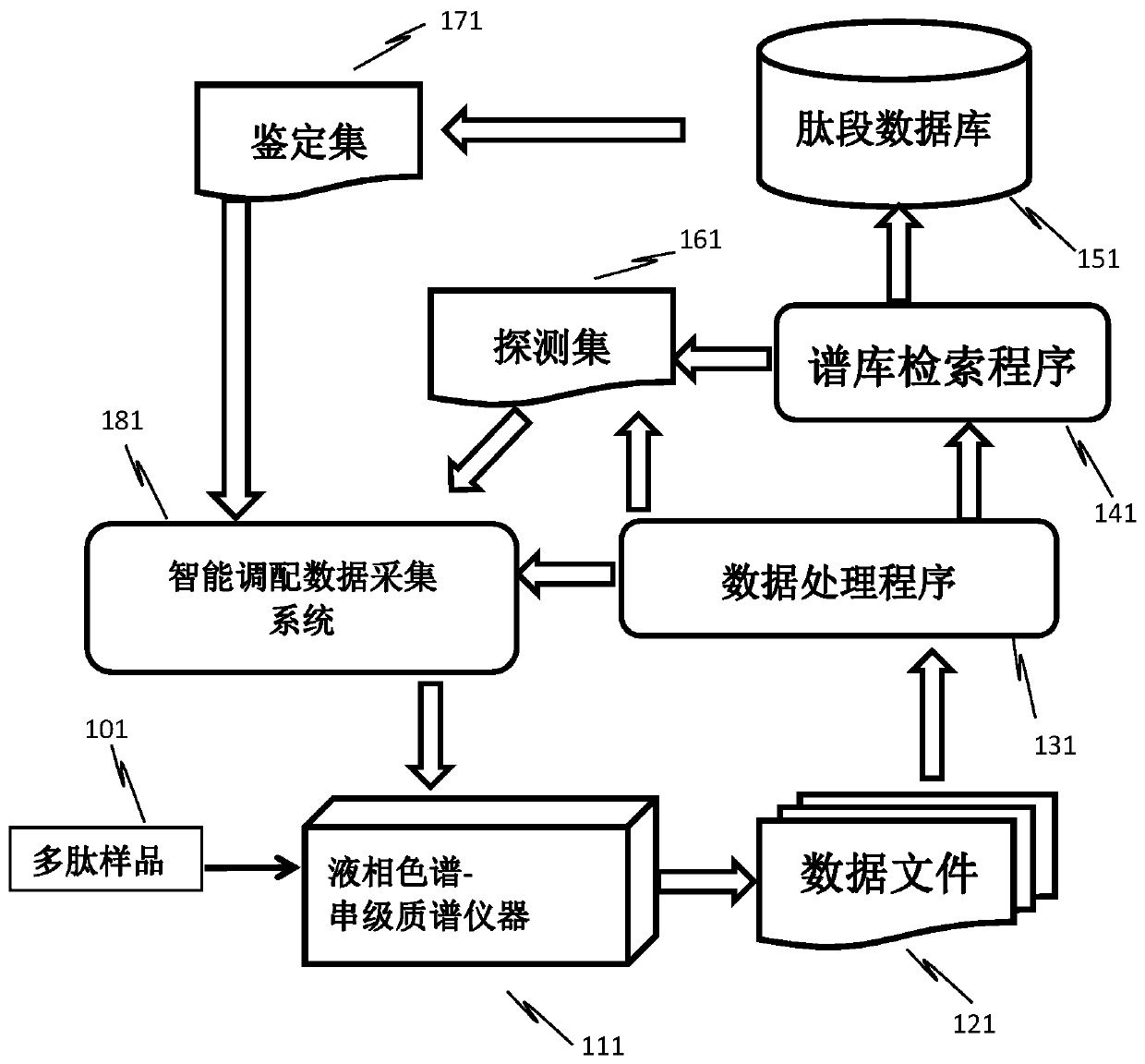
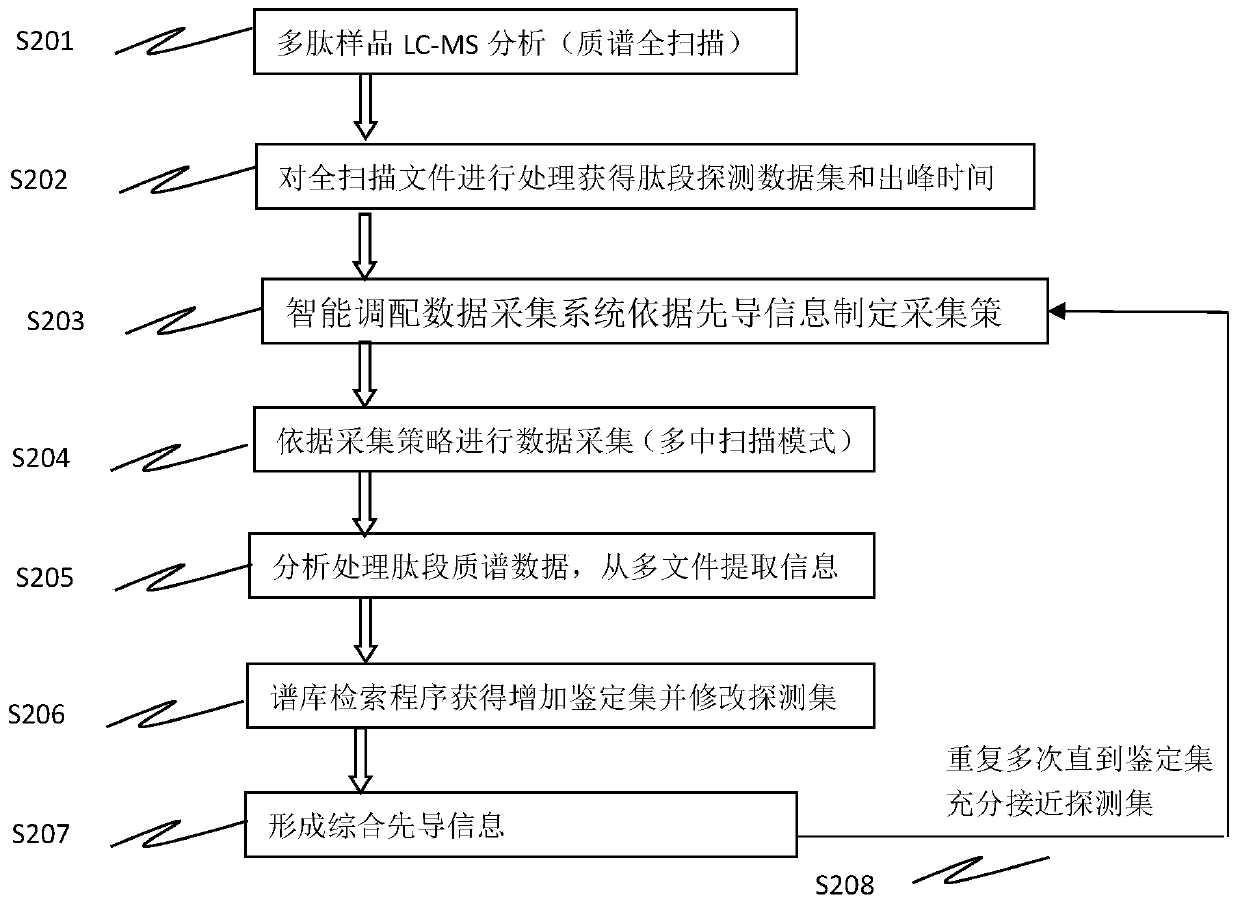
[0045] The first stage: extract the ones that need to be identified, and the number of identifications is 0, sort according to the peak height from large to small, and arrange the next identification according to the peak center time and scan time (3 MS / MS scans for each ion to be identified) Ion list, under the premise of ensuring the identification quality, arrange as many ions to be identified as possible in the same scan.
[0046] The second stage: extract the ones that need to be identified, the number of identifications is greater than 0, the identification mode ID is 3, sort according to the peak height from large to small, according to the peak center time and scan time (do 5 MS / MS scans for each ion to be identified Above) to arrange the next identification ion list, and arrange as many ions to be identified as possible in the same scan under the premise of ensuring the identification quality.
[0047]The third stage: Extract the ones that need to be identified, the n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com