Single-site DNA methylation detection kit
A detection kit and methylation technology, applied in the field of DNA methylation detection kits based on strand replacement probes and loop-mediated amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
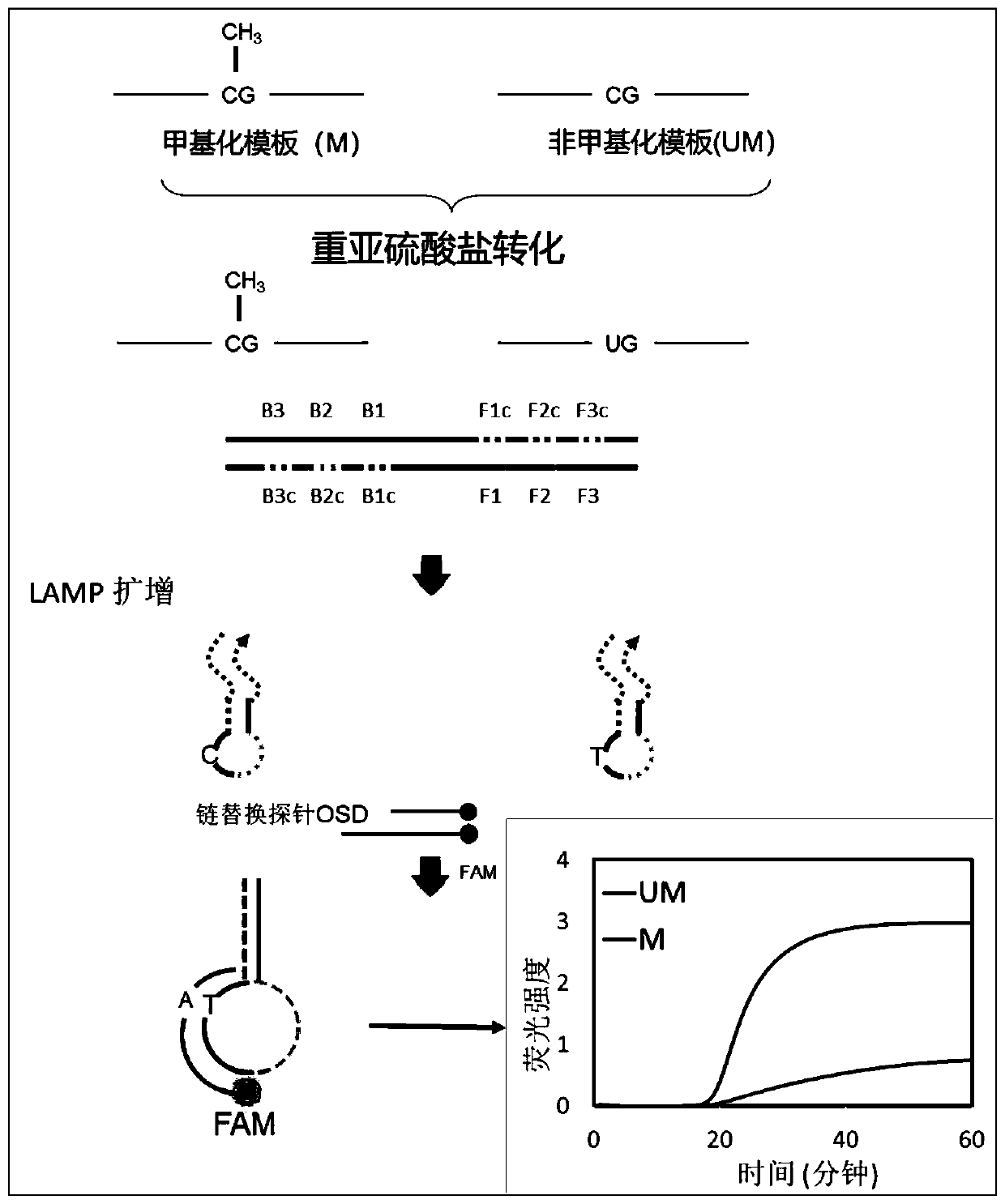
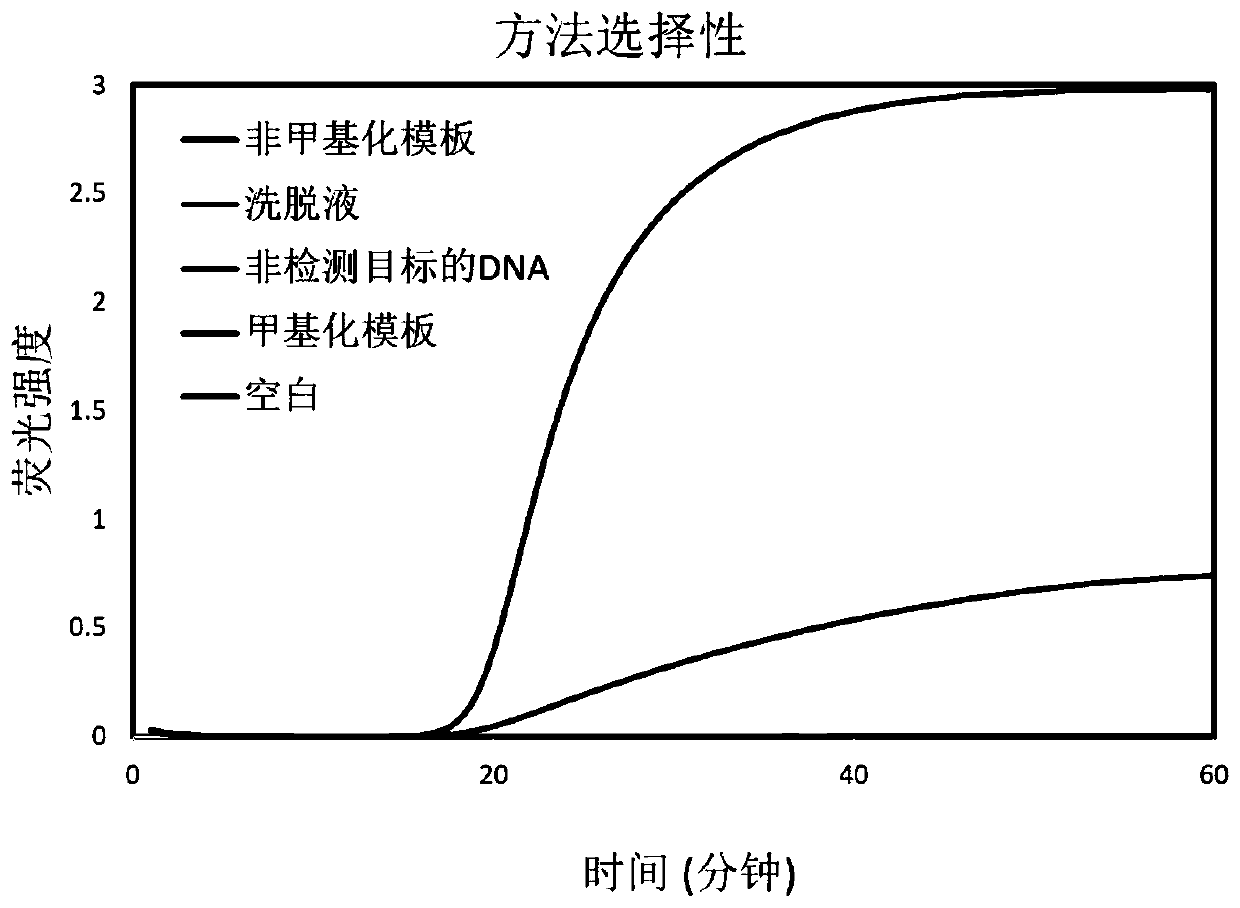
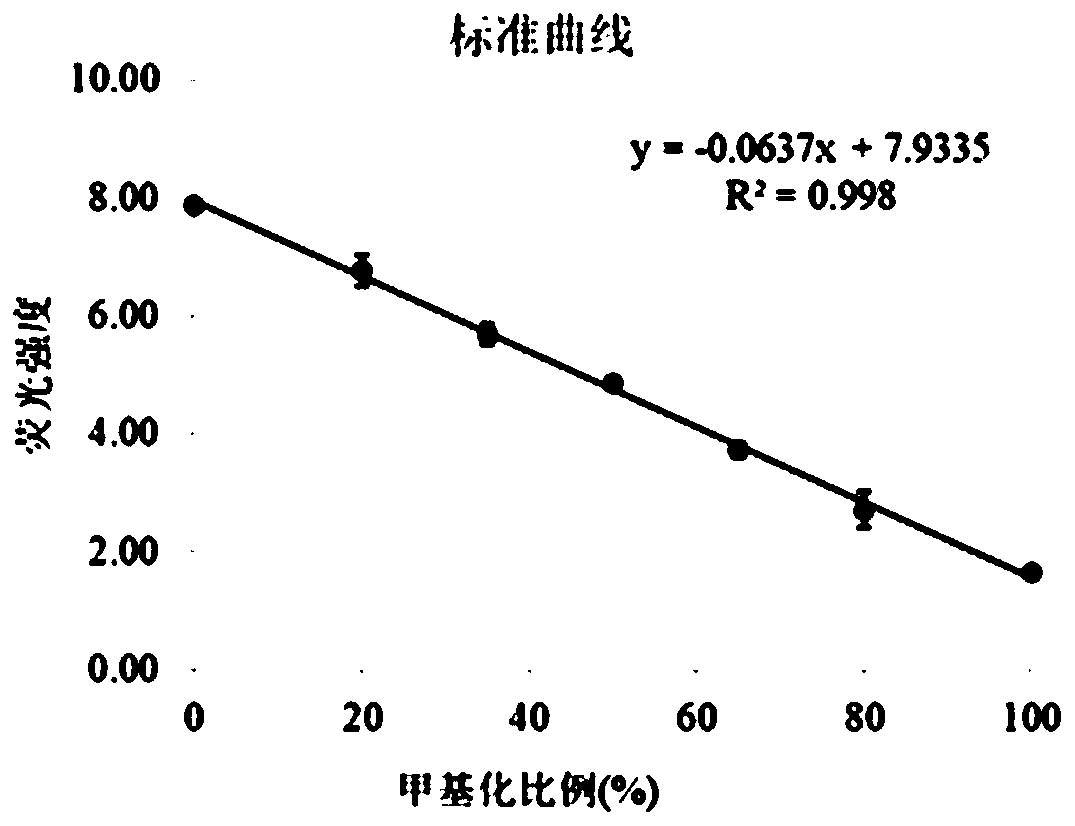
[0033] A single site DNA methylation detection kit, based on strand replacement probes and loop-mediated amplification to achieve DNA methylation detection, select the E-Box site in the proximal promoter region of the human genome SLC22A2 gene Methylation level locus detection object. The E-Box site sequence is designed to the position where the stem-loop structure of the LAMP amplification product is complementary to the F chain. Since the methylated template and the unmethylated template are treated with bisulfite, the bases are different (non-methylated The cytosine C is converted to uracil U, and the methylated cytosine remains unchanged). After LAMP amplification, a stem-loop structure product with a C / T single-base difference is produced. The single-base difference causes the OSD to be opened. The efficiency of the probe is different and the fluorescence intensity is different. Therefore, the LAMP-OSD method can detect the degree of methylation at the E-Box site in terms...
Embodiment 2
[0048] 1. Materials and methods
[0049] 1.1 Primers and probes
[0050] This kit uses Primer Premier 5 software to design PCR primers. Bisulfite conversion sequence primers are designed by MethPrimer (http: / / www.urogene.org / cgi-bin / methprimer / methprimer.cgi), and LAMP primers are designed by PrimerExplorer V5( http: / / primerexplorer.jp / lampv5e / index.html) design. The primer and probe sequences are as follows: Internal primer:
[0051] FIP(TATCAAAACCCTAATCCAAATTCTTGTTGGTTGGAGAATGAGT),
[0052] BIP(TGGTTTATATTACGTTGGGTTTTGTACGCACCGCCAACTT),
[0053] External primer:
[0054] F3(TTAGTATGTATTTYGAYGGT),
[0055] B3(CAACCAAAAACTTAATATACTCC),
[0056] Chain replacement detection probe OSD-F chain:
[0057] 5′-6-FAM-TTCTTCAAAACCCTATAAAAAAAACACACATATTTAACC-Inverted dT-3′
[0058] Chain replacement detection probe OSD-Q chain:
[0059] 5'-GTTTTTTTTATAGGGTTTTGAAGAA-BHQ1-3').
[0060] All DNA sequences were synthesized by Shanghai Shenggong Biological Engineering Co., Ltd.
[0061] 1.2 Bisulfite conversi...
Embodiment 3
[0115] A total of 30 tissue samples from kidney cancer patients were tested and verified by the LAMP-OSD method of this kit. The test results are as follows: Hypomethylation ( <30%) 28 copies, the positive result of the test is in full agreement with the reported result.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com