Metagenome absolute quantitation method
A genome, unit mass technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of large limitations of the bacterial flora analysis method, save the experimental cost and time, the experimental cost is low, and the operation is simple. easy effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0014] The following examples can enable those skilled in the art to understand the present invention more fully, but do not limit the present invention in any way.
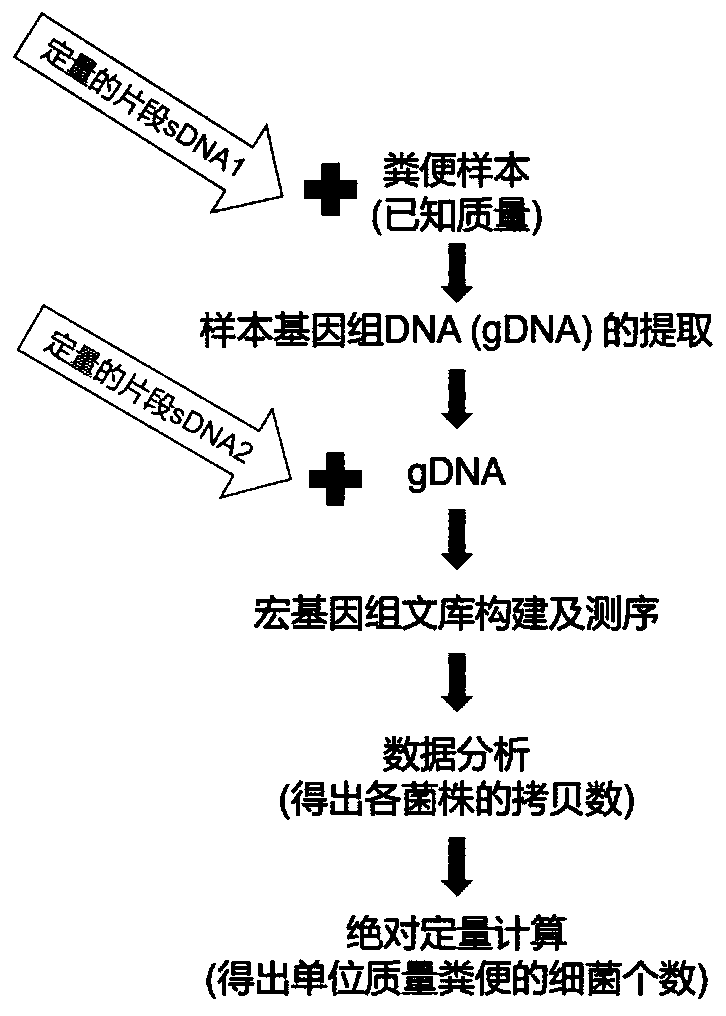
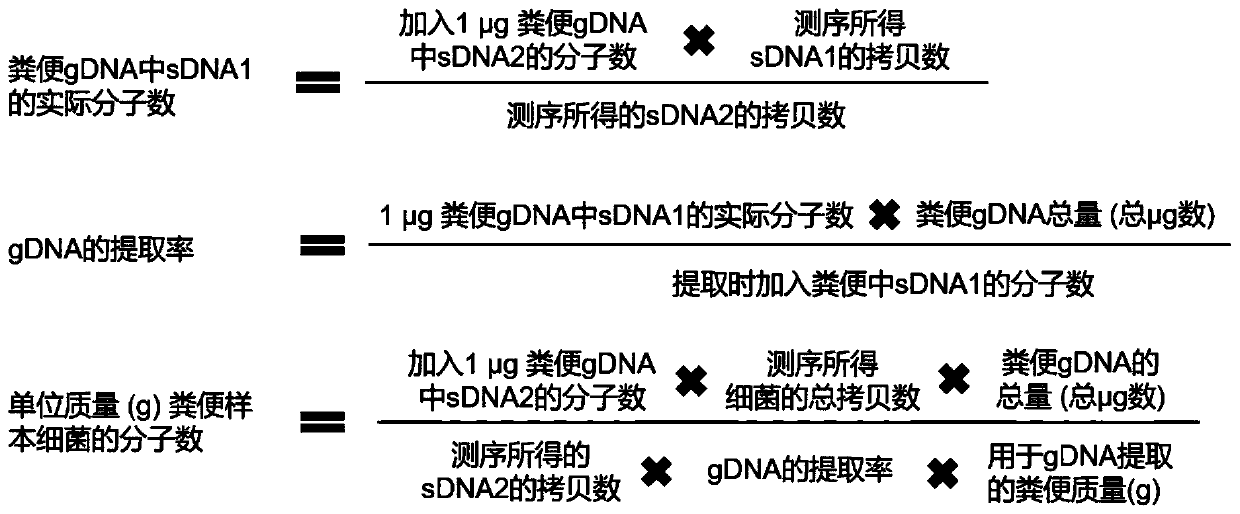
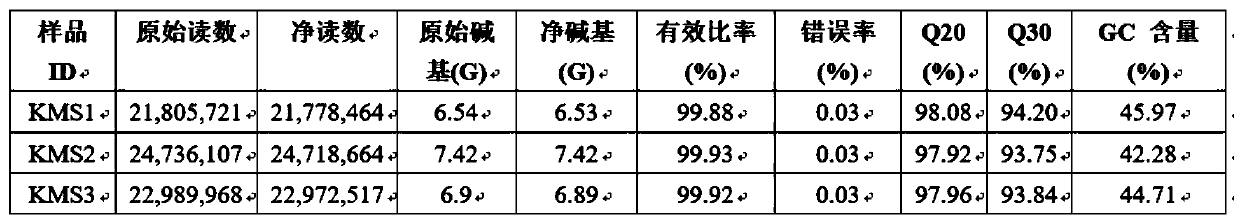
[0015] In order to improve the accuracy of the absolute quantitative results of target strains in stool samples, for example, one method is to use flow cytometry to detect the number of bacterial cells in stool samples, and then combine the intestinal flora abundance data obtained by 16S sequencing, Realize the absolute quantification of the bacterial flora; another method is to add a certain amount of external standard sequence to the sample DNA to construct and sequence the 16S amplicon library, and then according to the 16S amplicon readings of the external standard sequence and their absolute A standard curve was drawn for the copy number, and the absolute copy number of the 16S rRNA gene of the species corresponding to the operational taxonomic unit (OTU) representative sequence in the sample was calculated. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com