Method for breeding soybean mosaic virus (SMV) resistant soybeans
A soybean mosaic virus and soybean technology, applied in the biological field, can solve problems such as improvement of resistance to soybean mosaic virus disease that are not involved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
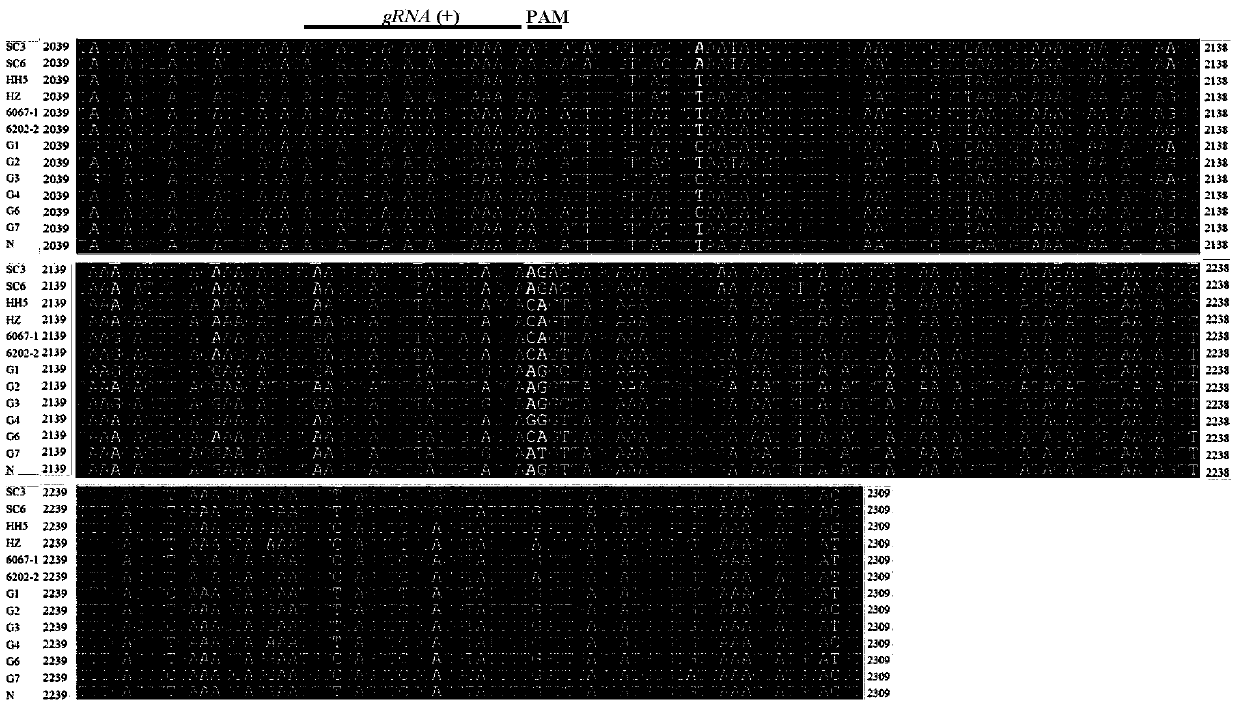
[0047] Embodiment 1, SMV genome multiple nucleotide sequence alignment and determination of gRNA target
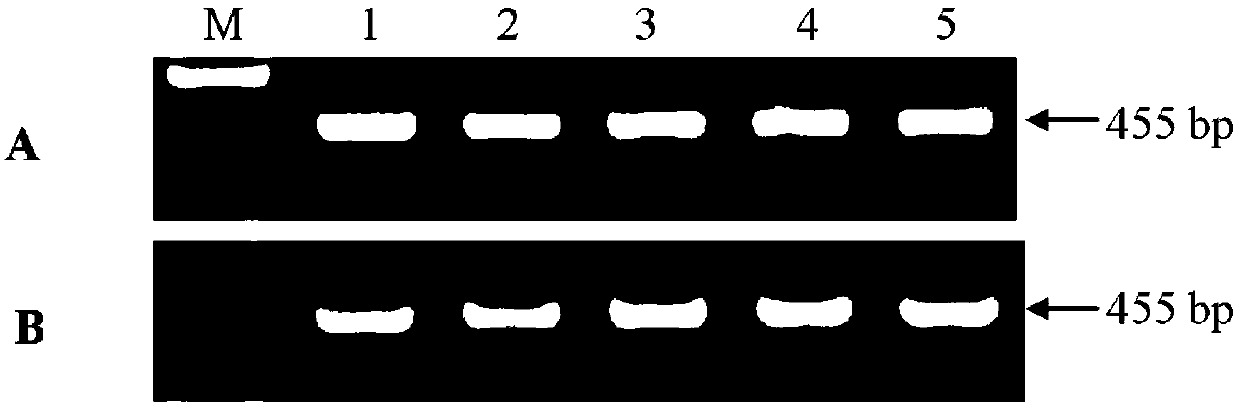
[0048] 1. Search for the full genomes of 13 SMV epidemic strains at home and abroad (the names and corresponding Genebank accession numbers of the 13 SMV epidemic strains are shown in Table 2) on the website of the National Center for Biotechnology Information (NCBI) sequence, and then utilize ClustalX 1.83 software to carry out multiple nucleotide sequence homology comparisons to the genomes of the SMV sense strand and the negative sense strand, and determine that the conserved interval of the SMVHC-Pro gene is 2039-2309nt (see figure 1 ) and 8373-8530nt (see figure 2 ), and then searched for the PAM site and determined the gRNA(+) target sequence with a similarity of 100% (see figure 1 ) and gRNA(-) target sequence (see figure 2 ).
[0049] gRNA (+) target sequence: 5'-AGGATGTACATAGCTAAAGA-3' (sequence 1 in the sequence listing).
[0050] gRNA (-) target sequence: ...
Embodiment 2
[0054] Example 2, T 1 Obtaining transgenic soybean and identification of SMV resistance at phenotype level, RNA level and protein level
[0055] 1. Obtaining recombinant Agrobacterium
[0056] 1. Take Microtube, add 5 μL HCss(+)-F aqueous solution (concentration is 10 μM), 5 μL HCss(+)-R aqueous solution (concentration is 10 μM) and 15 μL Autoclaved ddH 2 O, a mixed solution was obtained.
[0057] 2. After completing step 1, place the Microtube in a metal bath for annealing to obtain a reaction solution. The reaction liquid contains oligo dimer.
[0058] Annealing procedure: 95°C for 3 minutes, slowly cool at 95°C to 25°C (decrease about 3°C per minute), and 16°C for 5 minutes.
[0059] 3. After completing step 2, take another new Microtube, add 1 μL of the reaction solution obtained in step (2), 1 μL of Cas9 / gRNA Vector, 1 μL of Solution 1, 1 μL of Solution 2 and 6 μL of Autoclaved ddH 2 O, mix well.
[0060] 4. After step 3 is completed, the Microtube is placed in a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com