Application of single nucleotide polymorphism site in auxiliary identification of resistance of soybean mosaic virus
A single nucleotide polymorphism, soybean mosaic virus technology, applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem that gene functions are not fully elucidated
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036]Example 1 Excavation of SNP sites in soybean gene Glyma.07G250300
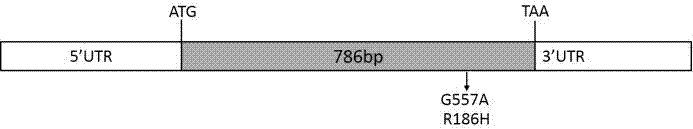
[0037] Glyma.07G250300 in the Soybean Genome Database (url ) in the corresponding gene is GmZFP3 (See Chinese patent ZL201110233787.2), without introns, resequenced by Zhou et al. 302 soybean materials were found to contain a SNP site in the open reading frame (ORF) of Glyma.07G250300, in which there are two types of G / A at the 557th nucleotide position, and it is a sense mutation, and the amino acid is represented by R (arginine) to H(histidine) (as in figure 1 ).
Embodiment 2
[0038] Example 2 Analysis of the Glyma.07G250300 Gene Coding Sequence in Soybean Materials
[0039] The tested soybean materials are: Willimas82 (corresponding sequence is GmZFP3); wild soybean BB52 (GsZFP3), SA001, SA002, SA025, SA062, SA101, SA162, SA189, SA242 and Willimas82.
[0040] The above 10 kinds of soybeans ( Glycine max (L.) Merr.) The genomic DNA in the leaves of the seedlings of the species; using the DNA as a template, PCR amplification was performed with a primer pair, and the primer sequence (used to amplify the open reading frame of Glyma.07G250300) was:
[0041] The forward primer (F) is shown in SEQ ID NO.1, and the reverse primer is shown in SEQ ID NO.2;
[0042] PCR reaction system: 2×Phanta Max Buffer 25 μL; dNTP Mix (10mM each) 1 μL; upstream and downstream primers (F / R) 4 μL (10 μM); Phanta Max Super-Fidelity DNA Polymerase (1 U / μL); template DNA 4 μL ( 20ng / μL); ddH 2 O to make up to 50 μL.
[0043] The PCR amplification program was pre-denaturat...
Embodiment 3
[0046] Example 3 Mosaic Virus Resistance Identification of Soybean Materials
[0047] The soybean materials listed in Table 1 were inoculated and identified by using two virus strains SC3 and SC7 (provided by the Soybean Research Institute of Nanjing Agricultural University) that are prevalent in the south. The identification methods and evaluation indicators (resistance and sensitivity) refer to Nanjing Agricultural University in 2000 Wang Xiuqiang's master's degree thesis "Research on Soybean Mosaic Virus (SMV) Strain Identification, Resistance Source Screening and Resistance Genetics in Huanghuai and the Middle and Lower Reaches of the Yangtze River".
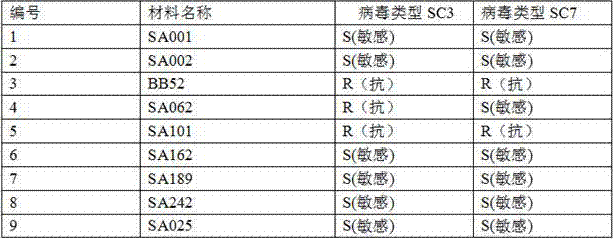
[0048] Table 1 Identification materials and results of soybean mosaic virus
[0049]
[0050] The results showed that BB52 and SA101 were resistant to both viruses, while SA062 was only resistant to SC3 and sensitive to SC7; while the rest of the materials were sensitive to both viruses (Table 1). Combined with the resul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com