Method capable of accurately detecting DNA viruses in human genome
A genome and human detection technology, applied in the fields of genomics, biochemical equipment and methods, proteomics, etc., can solve the problems of inability to cover HPV type detection, inability to determine whether HPV is integrated and integration status detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] An embodiment of the method for accurately detecting DNA viruses in the human genome of the present invention comprises the following steps:
[0092] For patient A with mild cervicitis, part of the cervical tissue was taken for capture sequencing. The data obtained by sequencing were analyzed as follows.
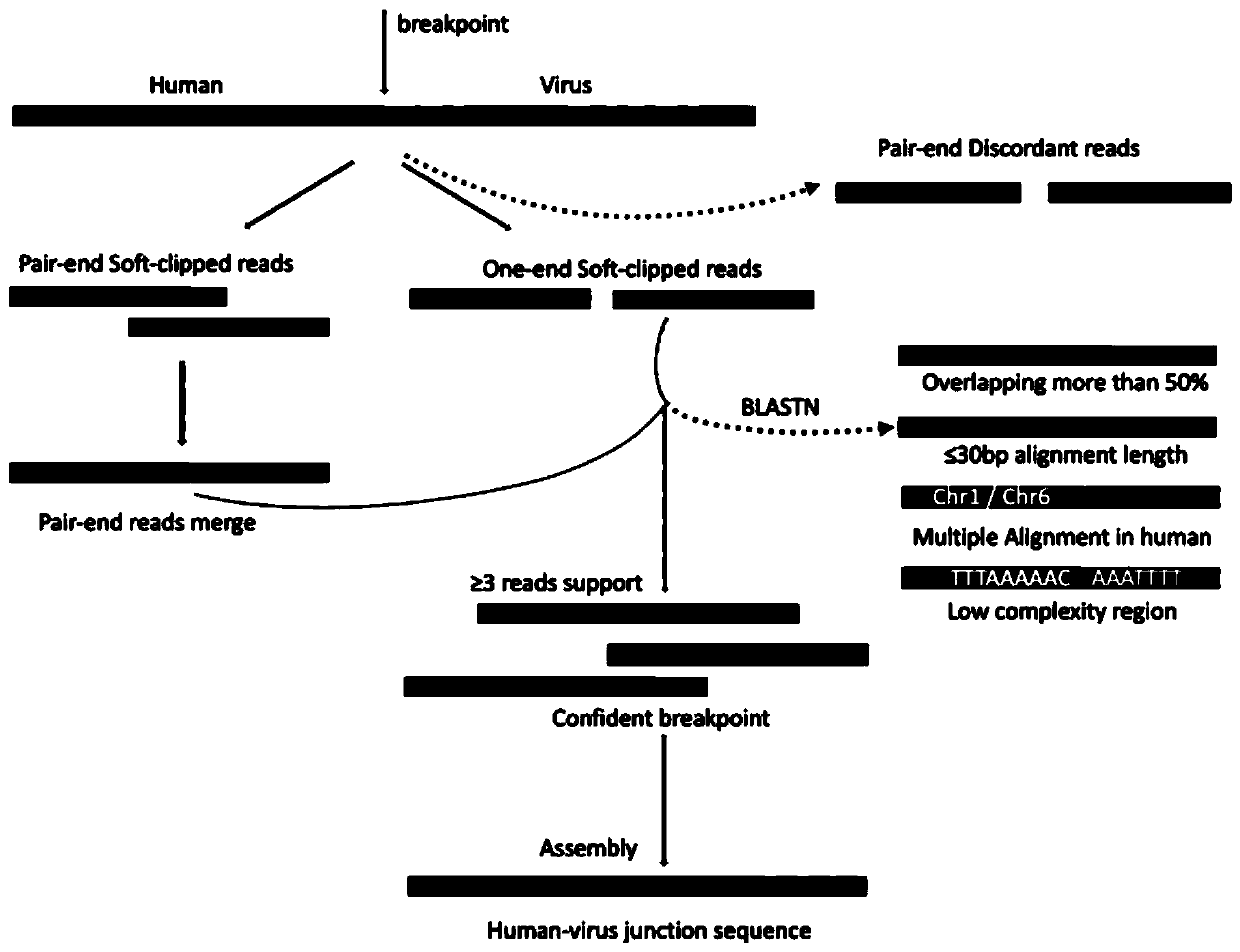
[0093] By filtering the comparison information of the sequencing reads, the reads from the viral DNA are accurately selected, and by removing the repeat offsets that may be caused during the library construction process, the reads of different types of viral DNA The number of segments is counted to indirectly reflect the load of the infected virus type. The specific steps are as follows:
[0094] (1) In the initial comparison process, all types of HPV virus genomes collected from the Papillomavirus Genome Database PaVE, the collected HPV virus genomes are used as pseudochromosomes, and are merged with the chromosomes of the human genome to construct a hybrid Genome;...
Embodiment 2
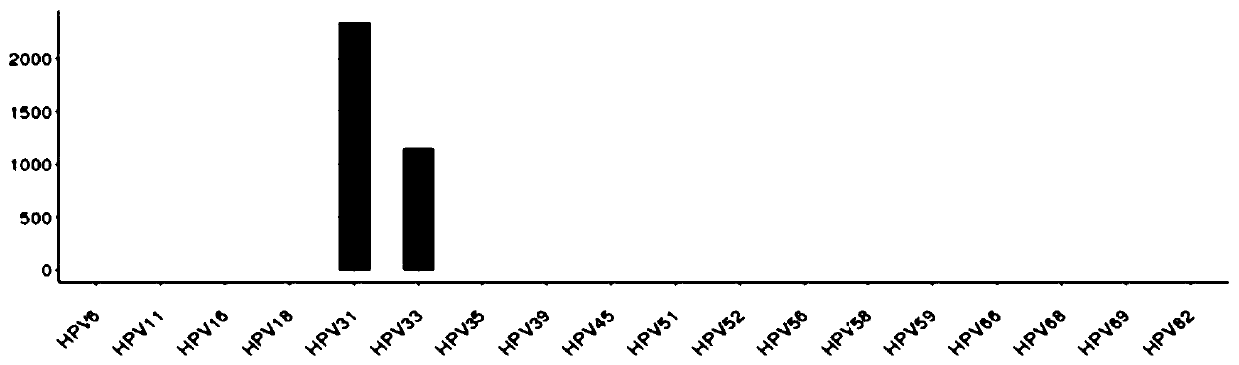
[0117] In mild cervicitis patient B (see Example 1 for the detection method), it was found that three HPV types (HPV16, HPV 31, HPV56) were infected (such as Figure 4 As shown), but no HPV integration was found, follow-up can be continued to avoid unnecessary colposcopy, which is different from the international guidelines for HPV16 positive recommendation for colposcopy referral.
Embodiment 3
[0119] In the follow-up follow-up of patient C with low-grade cervical lesions (see Example 1 for the detection method), it was found that high-risk HPV16 infection (such as Figure 5 As shown), but no HPV integration was found, follow-up can be continued to avoid unnecessary colposcopy, which is different from the international guidelines for HPV16 positive recommendation for colposcopy referral.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com