application of NTS/dNTS combination in preparation of plant mutants
A kind of technology of plants and variants, applied in the field of application in the preparation of plant mutants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Example 1, Construction of TS / dTS, TS / dNTS and NTS / dNTS Corresponding Vectors and Their Application in Rice Gene Replacement
[0098] 1. Construction of recombinant expression vector and description of replacement principle
[0099] 1. Construction of recombinant expression vector
[0100] Artificially synthesize the following recombinant expression vectors, each of which is a circular plasmid:
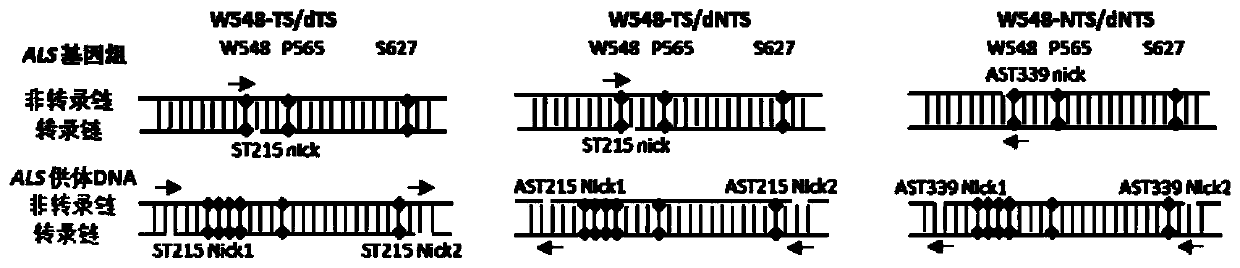
[0101] For the W548 site, there are three vectors, namely W548-TS / dTS, W548-TS / dNTS, and W548-NTS / dNTS.
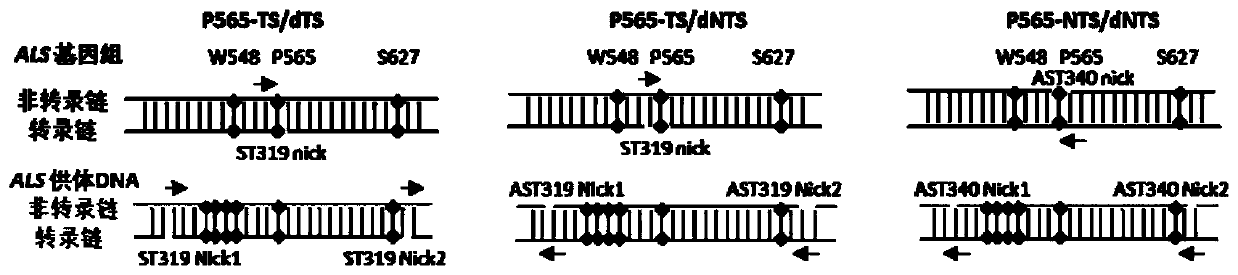
[0102] For the P565 site, there are three vectors, namely P565-TS / dTS, P565-TS / dNTS, and P565-NTS / dNTS.
[0103] Schematic diagram of the backbone structure of the recombinant expression vector figure 1 shown. The specific structure is described as follows:
[0104] The sequence of the W548-TS / dTS recombinant expression vector is sequence 1 in the sequence list. The 131-467th position of sequence 1 is the OsU3 promoter sequence, the 474-550th position is the tRNA sequ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com