Mass spectrum database establishing method based on virus identification
A technology for establishing a method and database, which is applied in the field of mass spectrometry database establishment based on virus identification, and achieves the effects of high sensitivity, high throughput and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
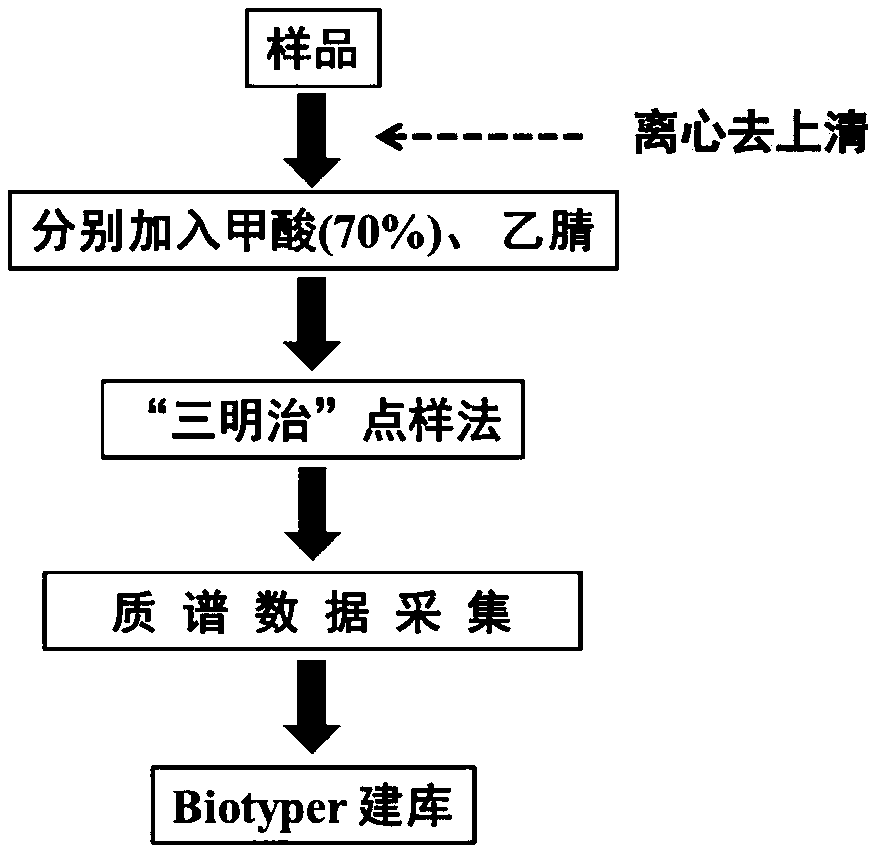
[0026] Virus sample preprocessing and database establishment
[0027] Virus sample pretreatment process:
[0028] Take 5 μL of the virus sample, centrifuge at 10,000 rpm at 0°C for 10 min, and then remove the supernatant. Add 2 μL of 10% trifluoroacetic acid solution to the pellet to fully dissolve the virus particles, then add 2 μL of acetonitrile and vortex to mix well. The obtained virus samples are pre-treated samples.
[0029] Virus mass spectrum acquisition:
[0030] First weigh 5 mg of CHCA and dissolve it in 1 mL of acetonitrile / water (3:2 volume ratio, containing 1% trifluoroacetic acid by volume) mixed solvent to obtain a 7 mg / mL matrix stock solution.
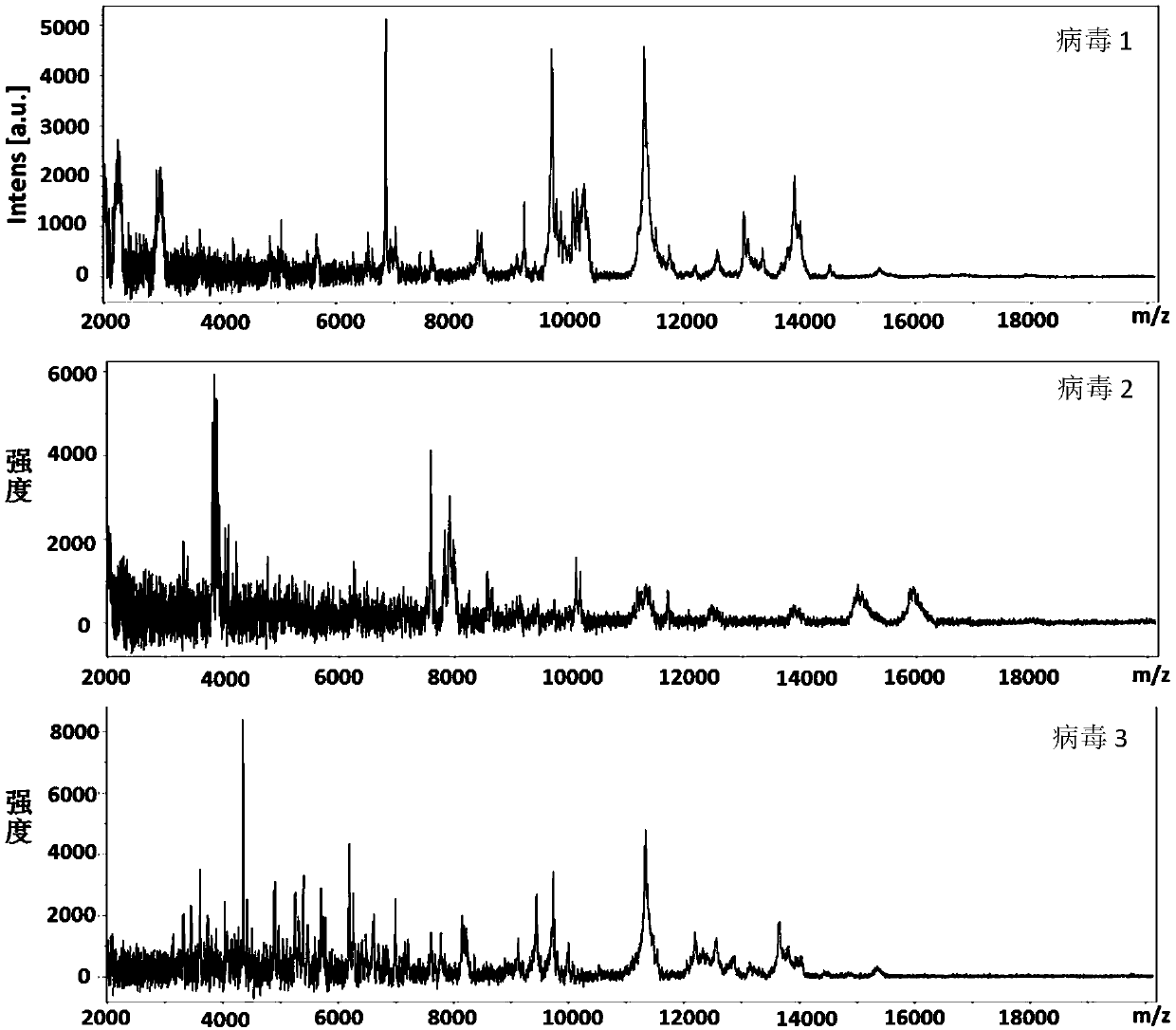
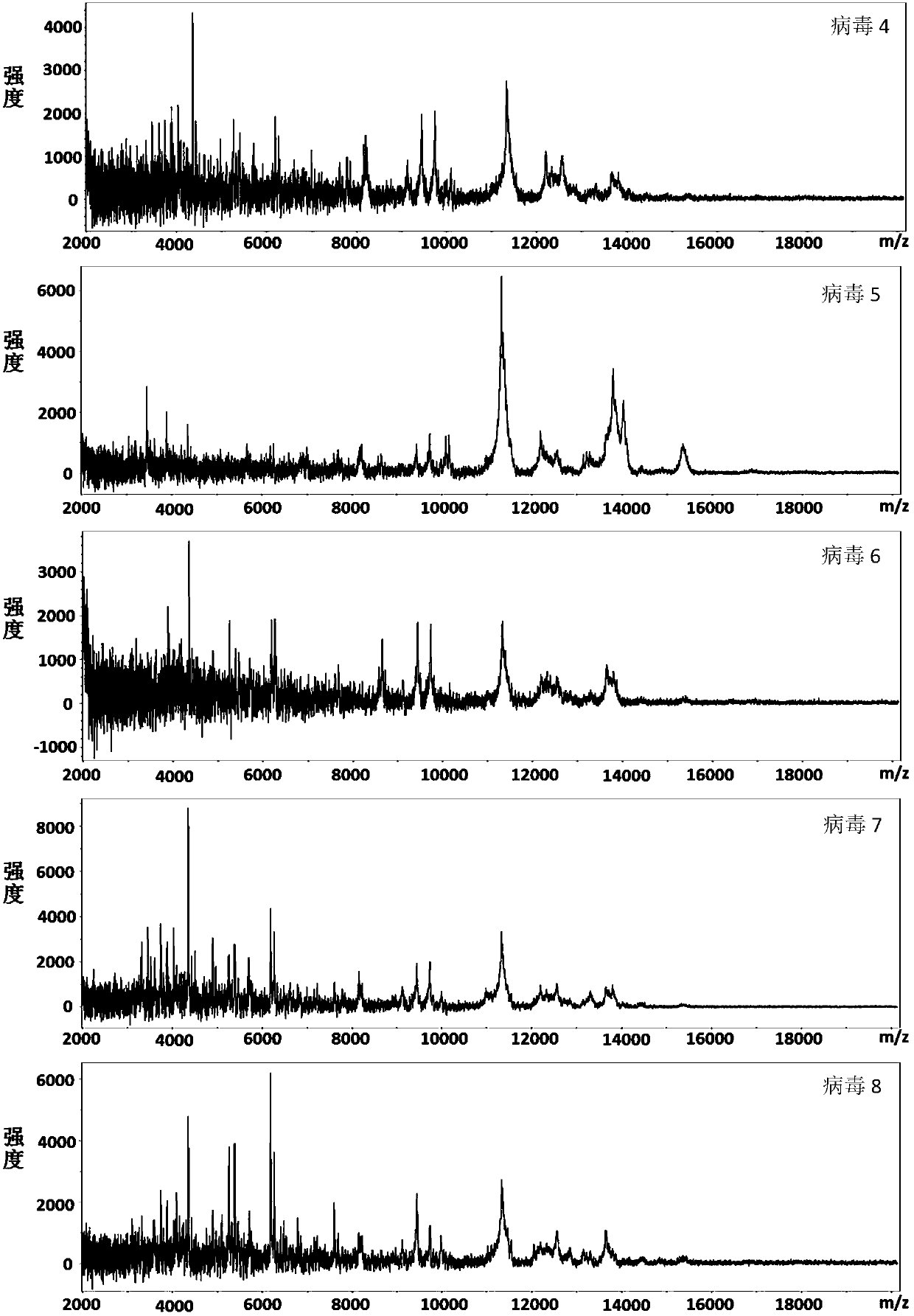
[0031] Add 1 μL of sample solution dropwise on the MALDI target plate, after drying, add 1 μL of CHCA matrix dropwise, and then spot 8 groups of samples in parallel. After all the solutions were dried, 24 (8×3) virus mass spectra were collected. The MALDI-TOF MS parameters during acquisition are set as follows: ...
Embodiment 2
[0036] MALDI-TOF MS detection of virus samples
[0037] Virus sample pretreatment process:
[0038] Take 8 μL of virus samples, centrifuge at 20,000 rpm at 4°C for 30 min, and remove the supernatant. Add 5 μL of 30% trifluoroacetic acid solution to the pellet to fully dissolve the virus particles, then add 5 μL of acetonitrile, and vortex to mix well. The obtained virus samples are pre-treated samples.
[0039] Virus mass spectrum collection:
[0040] First weigh 10 mg DHB and dissolve it in 2 mL of acetonitrile / water (3:2 volume ratio, containing 1% trifluoroacetic acid by volume) mixed solvent to obtain a 5 mg / mL matrix stock solution.
[0041] Add 1 μL of sample solution dropwise on the MALDI target plate, after drying, add 1 μL of LDHB matrix dropwise, and then spot 8 groups of samples in parallel. After all the solutions were dried, 24 (8×3) virus mass spectra were collected. The MALDI-TOF MS parameters during acquisition are set as follows: laser energy (60%), frequ...
Embodiment 3
[0046] MALDI-TOF MS detection of virus samples
[0047] Virus sample pretreatment process:
[0048] Take 10 μL of the virus sample, centrifuge at 10° C. at 20,000 rpm for 45 minutes, and remove the supernatant. Add 5 μL of 50% trifluoroacetic acid solution to the precipitate to fully dissolve the virus particles, then add 5 μL of acetonitrile, and vortex well. The obtained virus samples are pre-treated samples.
[0049] Virus mass spectrum collection:
[0050] First weigh 20 mg of SA and dissolve it in 1 mL of acetonitrile / water (3:2 volume ratio, containing 0.1% trifluoroacetic acid by volume) mixed solvent to obtain a 20 mg / mL matrix stock solution.
[0051] Add 1 μL of sample solution dropwise on the MALDI target plate, after drying, add 1 μL of SA matrix dropwise, and then spot 8 groups of samples in parallel. After all the solutions were dried, 24 (8×3) virus mass spectra were collected. The MALDI-TOF MS parameters during acquisition are set as follows: laser energy ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com