Primer group and crude drug identification method using primer group
A primer set and crude drug technology, applied in biochemical equipment and methods, microbial measurement/inspection, recombinant DNA technology, etc., can solve the problems of inability to accurately identify the original plant and difficulty in determining the base sequence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0241]
[0242] 1 Introduction
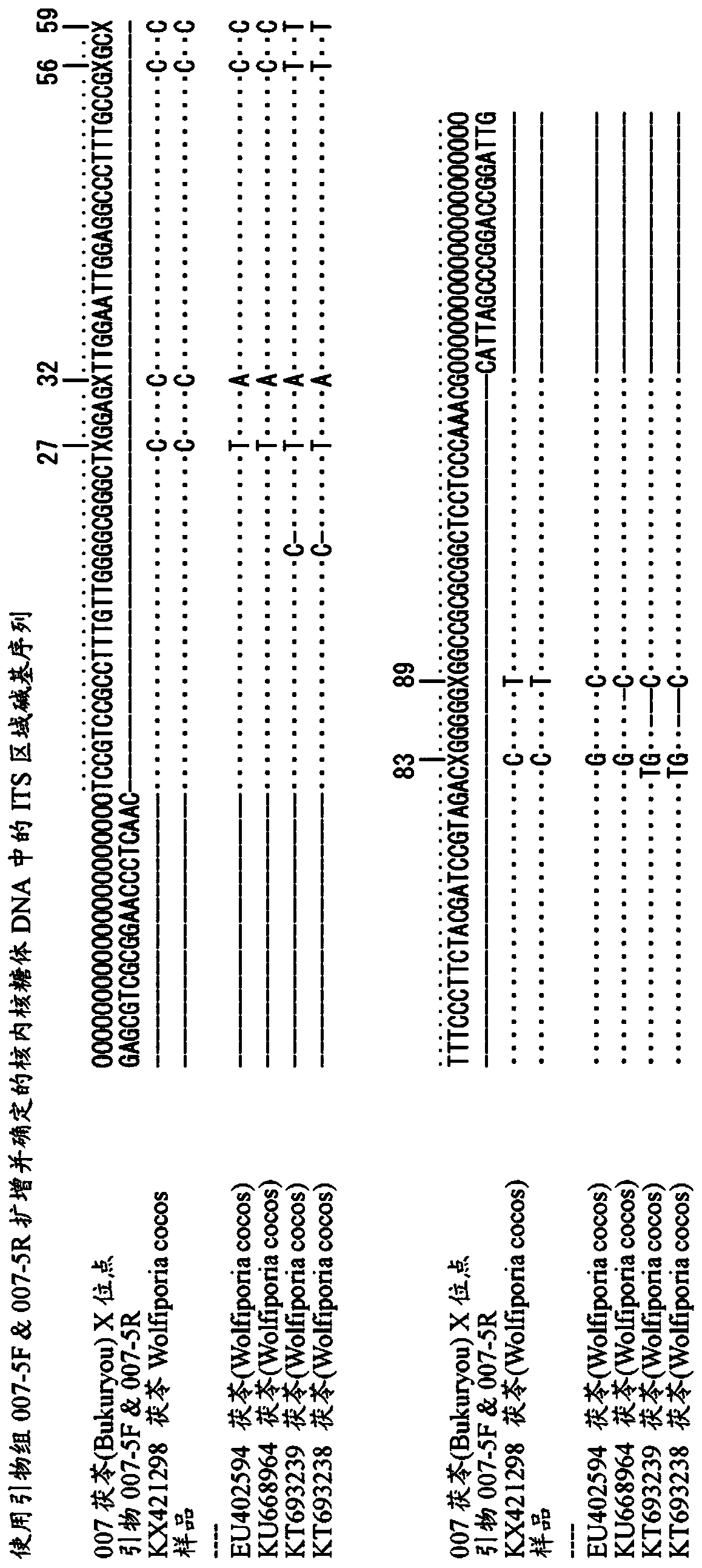
[0243] In the seventeenth revised edition of the Japanese Pharmacopoeia, the original plant of Poria cocos is Wolfiporia cocos Ryvarden et Gilbertson (Poria cocos Wolf) (Poria cocos Wolf) (Poria cocos Wolf). However, it is well known that there are many kinds of internal mutations in the same species. Although there are varieties in the Japanese market that meet the regulations of the Japanese Pharmacopoeia, it is also possible to circulate multiple lines with different base sequences. The ingredients contained in different strains may be different. In order to ensure the quality of the medicine, it is an effective way to avoid mixing these different strains.
[0244] Therefore, the present inventors conducted a comprehensive study on the genomic DNA of various Poria strains and their related bacteria. The results showed that in the sequence of the ITS region in the ribosomal DNA, Poria, a major circulating product in the market, has a fixed sequen...
Embodiment 2
[0285]
[0286] 1 Introduction
[0287] In the seventeenth revised edition of the Japanese Pharmacopoeia, the original plant of Rehmannia glutinosa is Rehmanniaglutinosa Liboschitz var.purpurea Makino or Rehmannia glutinosa Liboschitz (Scrophulariaceae). However, other closely related species such as Rehmannia henryi may be circulated in the market. In order to ensure the quality of drugs, the key is to avoid misuse and mixing them.
[0288] Therefore, the inventors conducted a comprehensive study on the genomic DNA or chloroplast DNA of Rehmannia glutinosa and its relatives. The results showed that in the sequence of ITS region in ribosomal DNA, Rehmannia glutinosa has a fixed sequence in this region, which can be distinguished from other related species. Therefore, this region is a useful DNA marker for distinguishing Rehmannia glutinosa from other related species.
[0289] However, it is often difficult to determine the sequence from crude drugs due to the following reasons: 1) ...
Embodiment 3
[0314]
[0315] 1 Introduction
[0316] In the seventeenth revised edition of the Japanese Pharmacopoeia, the original plant of Corydalis is Corydalisturtschaninovii Besser forma yanhusuo Y.H.Chou et C.C.Hsu (Papaveraceae). However, other related species, Corydalis ambigua, may be circulated in the market. In order to ensure the quality of the drug, the key is to avoid misuse and mixing them.
[0317] Therefore, the inventors conducted a comprehensive study on the genomic DNA or chloroplast DNA of Corydalis and its relatives. The results showed that in the sequence of the ITS region of ribosomal DNA, Corydalis has a fixed sequence in this region, which can be distinguished from other related species. Therefore, this region is a useful DNA marker for distinguishing Yanhusuo from other related species.
[0318] However, it is often difficult to determine the sequence from crude drugs due to the following reasons: 1) DNA damage and fragmentation caused by time changes and processing; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com