Amplifying and sequencing primer for subtyping and drug resistance detection of helicobacter pylori, and reagent kit
A technology of Helicobacter pylori and sequencing primers, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., which can solve the problems of multiple mutation types and incomplete detection of Helicobacter pylori, and avoid Complicated and difficult, reducing manpower and material resources, and the effect of multiple experimental information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
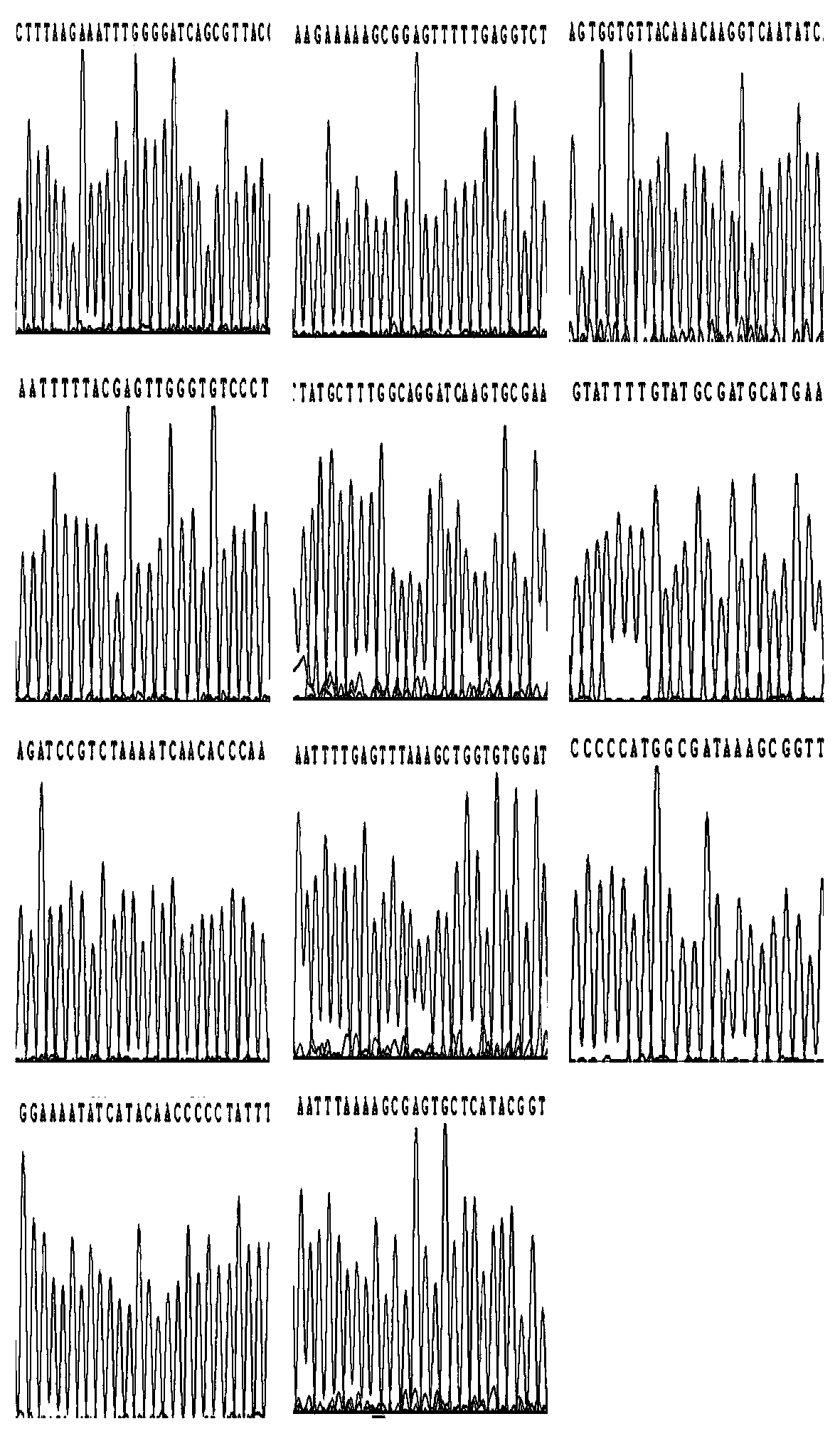
[0063] A set of specific primers for the amplification and sequencing of Helicobacter pylori, including the amplification and sequencing primers of UreA are SEQ ID NO.1 and SEQ ID NO.2, and the amplification and sequencing primers of 16S rRNA are SEQ ID NO. 3 and SEQ ID NO.4, rdxA amplification and sequencing primers are SEQ ID NO.5 and SEQ ID NO.6, 23S rRNA amplification and sequencing primers are SEQ ID NO.7 and SEQ ID NO.8, gyrA The amplification and sequencing primers are SEQ ID NO.9 and SEQ ID NO.10, the amplification and sequencing primers of PBP1 are SEQ ID NO.11 and SEQ ID NO.12, and the amplification and sequencing primers of cagA are SEQ ID NO.13 And SEQ ID NO.14, the amplification of vacAs and sequencing primer are SEQ ID NO.15 and SEQ ID NO.16, the amplification of vacAm and sequencing primer are SEQ ID NO.17 and SEQ ID NO.18, the amplification of vacAi The amplification and sequencing primers are SEQ ID NO.19 and SEQ ID NO.20, and the amplification and sequencing ...
Embodiment 2
[0065] A kit for amplifying and sequencing Helicobacter pylori, consisting of the following reagents:
[0066] Reagent A: Consists of 2mL buffer containing Green I dye, 0.2mM dNTPs (including dATP, dCTP, dGTP, dTTP), 80mM Tris-HCl, 80mM KCl, 40mM (NH4)2SO4, 6mM MgSO4, 100U TaqDNA polymerase;
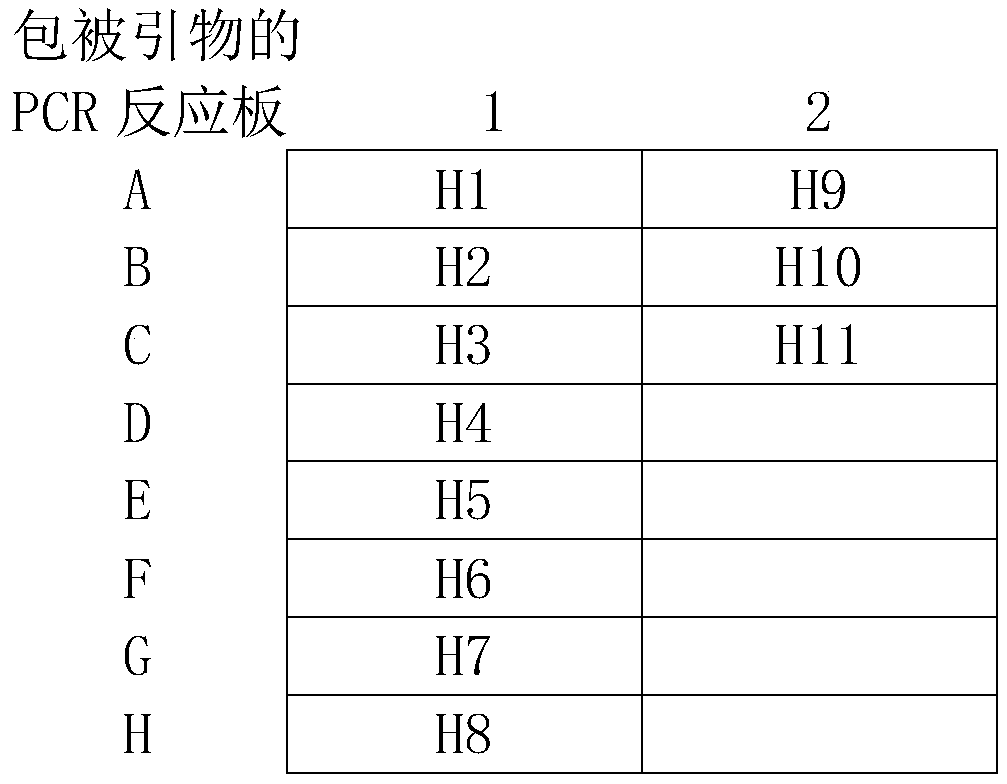
[0067] Reagent H1: 30 μL aqueous solution of sequencing primers targeting UreA, containing 4 μM SEQ ID NO.1;
[0068] Reagent H2: 30 μL aqueous solution of sequencing primers targeting 16S rRNA, containing 4 μM SEQ ID NO.3;
[0069] Reagent H3: 30 μL aqueous solution of sequencing primers for rdxA, containing 4 μM SEQ ID NO.5;
[0070] Reagent H4: 30 μL aqueous solution of sequencing primers targeting 23S rRNA, containing 4 μM SEQ ID NO.7;
[0071] Reagent H5: 30 μL aqueous solution of sequencing primers targeting gyrA, containing 4 μM SEQ ID NO.9;
[0072] Reagent H6: 30 μL aqueous solution of sequencing primers against PBP1, containing 4 μM SEQ ID NO.11;
[0073] Reagent H7: 30 ...
Embodiment 3
[0091] Real-time fluorescent quantitative PCR (real-time FQ PCR) is used to amplify all the sequences of Helicobacter pylori with the kit of the present invention. First, calculate the required amount of each reagent according to the number of experimental specimens. The experimental steps are as follows:
[0092] 1. Amplification reaction solution configuration: Prepare the reaction solution (1 test sample) in order, add 120 μL of water, 150 μL of reagent A, and 30 μL of DNA sample, a total of 300 μL; mix well.
[0093] 2. Add 25 μL of amplification reaction solution to each tube in sequence, and cover the caps.
[0094] 3. Amplify according to the PCR program designed in the experiment: pre-denaturation at 95°C for 3 minutes, denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, and extension at 72°C for 35 seconds. During the extension period, the fluorescence value was obtained for 35 cycles, and finally stored at 25°C. ℃; take it out in time for sequencing. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com