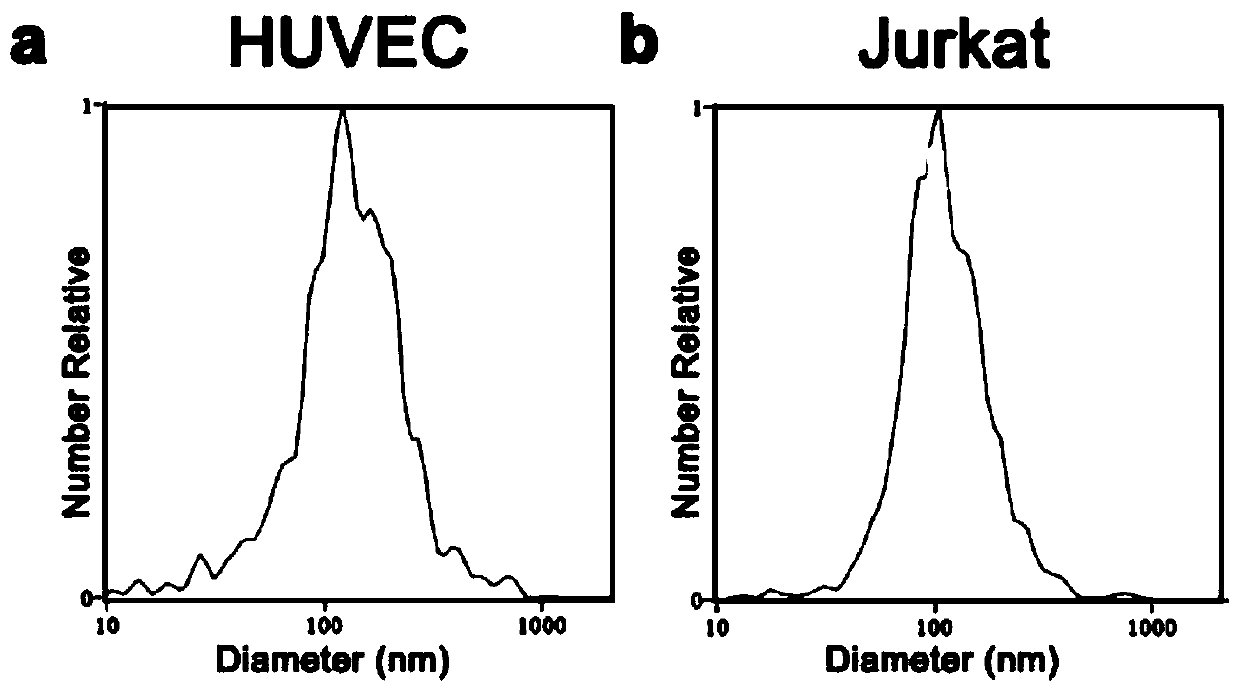
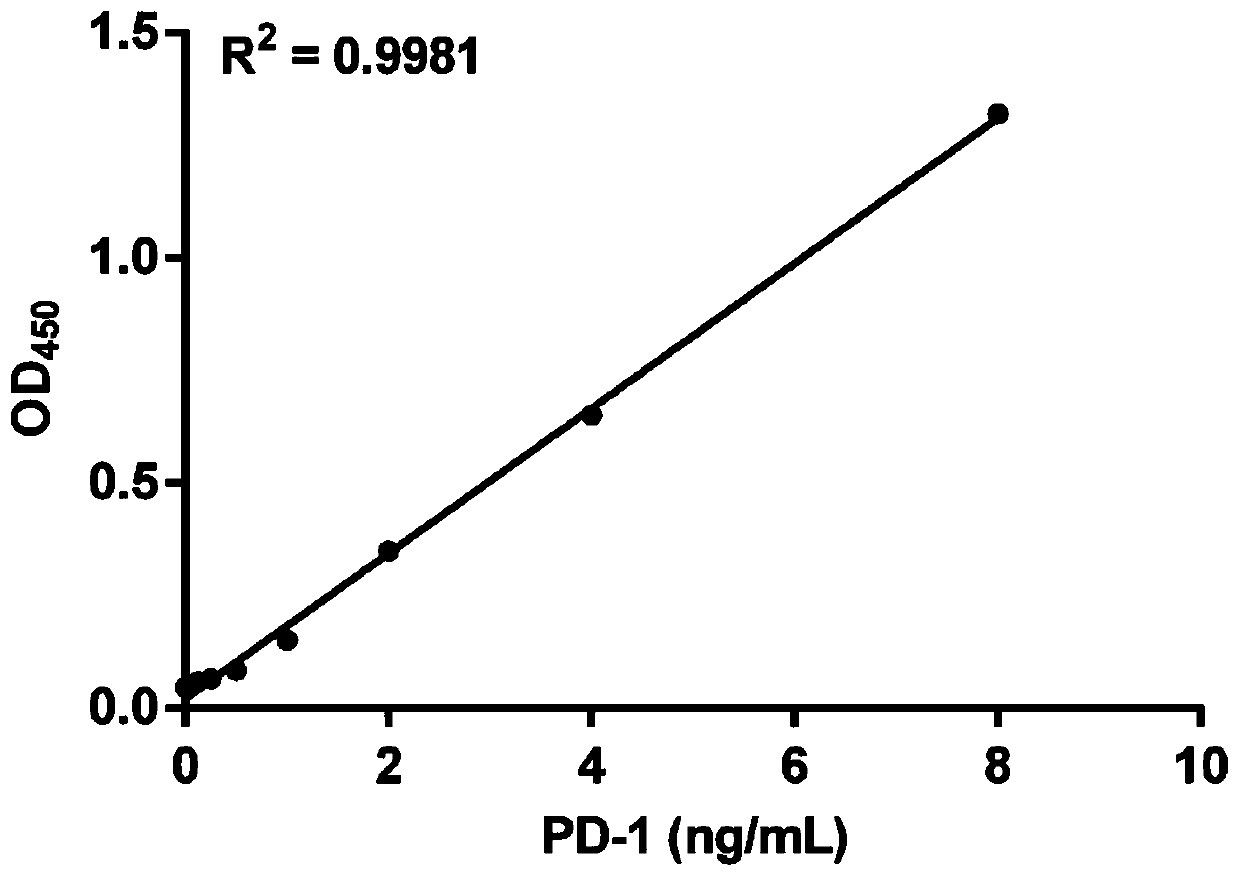
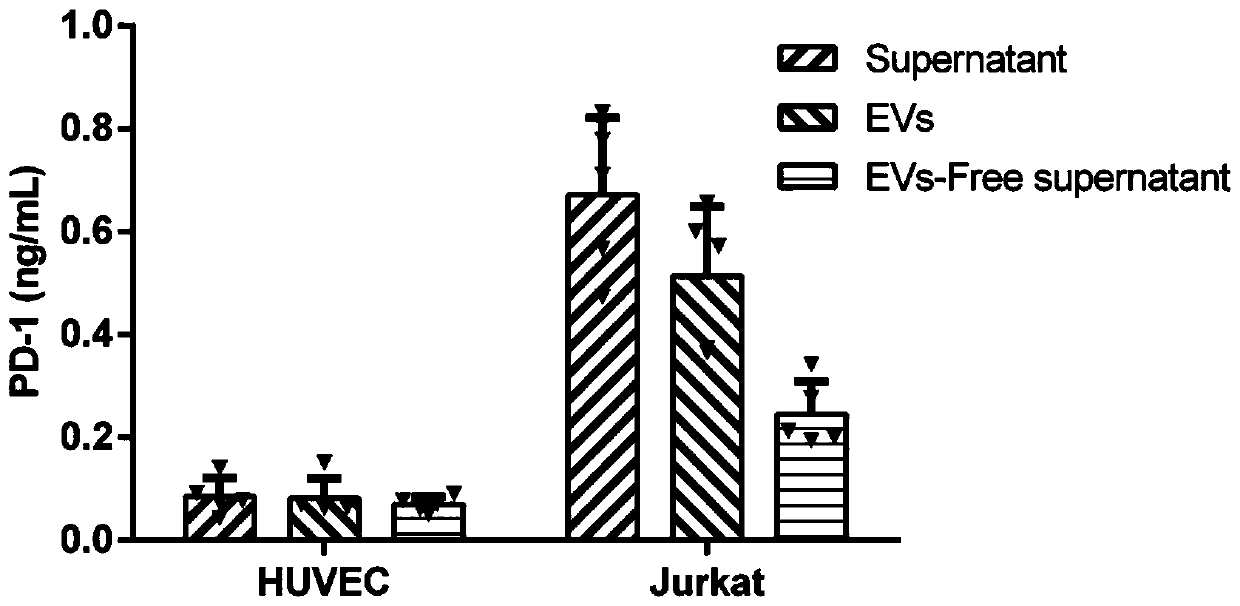
Method for efficiently and quantitatively detecting PD-1 level in extracellular vesicles, ELISA kit and use method
A PD-1, quantitative detection technology, applied in the field of molecular biology and biology, can solve the problems of unfavorable clinical application, lack of standardized operation procedures, unclear and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Construction of CD63-PD-1 fusion protein
[0045] 1. According to PubmedGene query to human CD63 gene sequence (Gene ID: 967), select the Ala103 to Val203 translation fragment; PD-1 gene sequence (Gene ID: 5133), select the Leu125 to Gln167 fragment. Design the His protein tag sequence, start codon, and stop codon; insert the connexin sequence between the two sequences
[0046] (GGTGGTGGTGGTAGCGGTGGTGGCGGTAGTGGTGGTGGTGGTAGC), introduce EcoR1, Xhol double restriction sites, design a pair of positive and negative primers (Table 1), and perform PCR amplification to obtain the CD63-PD-1 gene fragment. .
[0047] Table 1. CD63-PD-1PCR primers
[0048]
[0049] 2. Ligate the CD63-PD-1 gene fragment with the Pet28a plasmid that has been digested by Ecorl and Xhol through Exnase recombinase (Novazyme, Cat. No. C113-01), and obtain the recombinant plasmid CD63-PD after transformation and screening -1, its nucleic acid sequence is shown in SEQ ID No: 1, and its a...
Embodiment 2
[0056] Example 2: Construction of CD9-PD-1 fusion protein
[0057] 1. According to Pubmed Gene query to the human CD9 gene sequence (Gene ID: 928), select the Ser112 to Ile195 translation fragment; PD-1 gene sequence (Gene ID: 5133), select the Leu125 to Gln167 fragment. Design the His protein tag sequence, start codon, and stop codon; insert the connexin sequence between the two sequences
[0058] (GGTGGTGGTGGTAGCGGTGGTGGCGGTAGTGGTGGTGGTGGTAGC), introduce EcoR1, Xhol double restriction sites, design a pair of positive and negative primers (Table 2), and perform PCR amplification to obtain the CD3-PD-1 gene fragment.
[0059] Table 2 CD9-PD-1PCR primers
[0060]
[0061] 2. Ligate the CD9-PD-1 gene fragment with the Pet28a plasmid that has been digested by Ecorl and Xhol through Exnase recombinase (Novazyme, Cat. No. C113-01), and obtain the recombinant plasmid CD9-PD after transformation and screening -1, its nucleic acid sequence is shown in SEQ ID No: 2, and its amino ...
Embodiment 3
[0068] Example 3: Construction of CD81-PD-1 fusion protein
[0069] 1. According to the PubmedGene query of the human CD81 gene sequence (Gene ID: 975), select the translated fragment from Phe113 to Lys201; PD-1 gene sequence (Gene ID: 5133), select the fragment from Leu125 to Gln167. Design the His protein tag sequence, start codon, and stop codon; insert the connexin sequence between the two sequences
[0070] (GGTGGTGGTGGTAGCGGTGGTGGCGGTAGTGGTGGTGGTGGTAGC), introduce EcoR1, Xhol double restriction sites, design a pair of forward and reverse primers (Table 3), and perform PCR amplification to obtain the CD3-PD-1 gene fragment.
[0071] Table 3. CD81-PD-1PCR primers
[0072]
[0073] 2. Ligate the CD81-PD-1 gene fragment with the Pet28a plasmid that has been digested by Ecorl and Xhol through Exnase recombinase (Novazyme, Cat. No. C113-01), and obtain the recombinant plasmid CD81-PD after transformation and screening -1, its nucleic acid sequence is shown in SEQ ID No: 3...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com