Method for detecting circulating miRNA by using DNA machine based on quantum dot micelle spherical nucleic acid
A technology of quantum dots and DNA enzymes, applied in the field of biomedicine, can solve the problems of inconvenient functional nucleic acid integration, small particle size of water-soluble quantum dots, etc., and achieve the effect of high sensitivity and multivariate detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
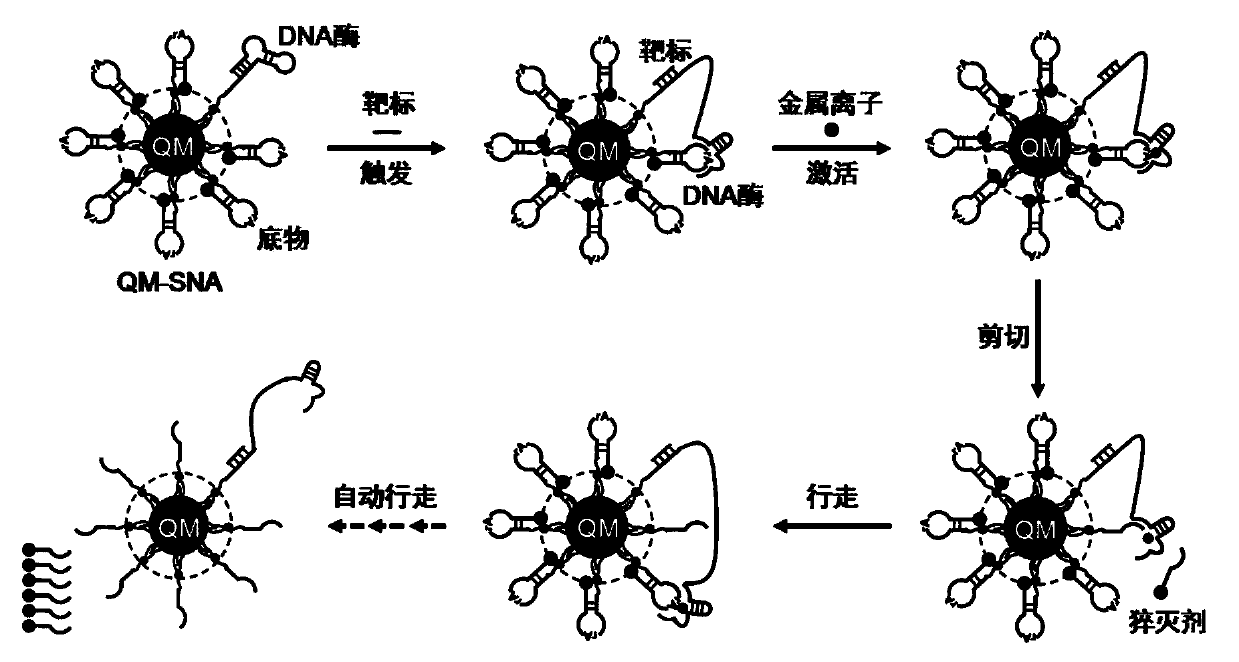
[0024] Sequence design of three targets, three DNases, and three substrates
[0025] Such as figure 1 As shown, the corresponding DNA enzymes (DNAzyme 1-3) and substrates (Substrate 1-3 ). The DNase sequence mainly includes three parts: the target binding region, the (substrate) track binding region, the catalytic center region, and the modified amino group at the 5' end for coupling to the QM surface. Among them, the target binding region sequence is the complementary sequence of the target, which is used to hybridize with the target to open the stem-loop structure of DNase; the orbital binding region includes two sequences of the upper and lower arms, which are used to bind to the substrate; the catalytic center region is used to bind metal ions Recognizes and cuts the rA site. The substrate sequence mainly includes the complementary sequence of the upper and lower arms, the rA site, the modified amino group at the 3' end is used for coupling to the QM surface, and the BH...
Embodiment 2
[0029] Preparation of three-color QM-SNA loaded with DNase and substrate
[0030] The three-color QM-SNA is prepared by chemical cross-linking, and the preparation process is the same. Take 50 μL of QM solution (concentration: 50 nM), 10 μL of EDC solution (100 μM) and Sulfo-NHS solution (50 μM) into 0.01M, pH7.4 phosphate buffer solution (total volume 500 μL), mix and stir for 15 min Afterwards, 25 μL of DNase solution (45 nM) and 225 μL of substrate nucleic acid solution (455 nM) were sequentially added, reacted for 4 hours at an appropriate stirring speed, and purified by ultrafiltration.
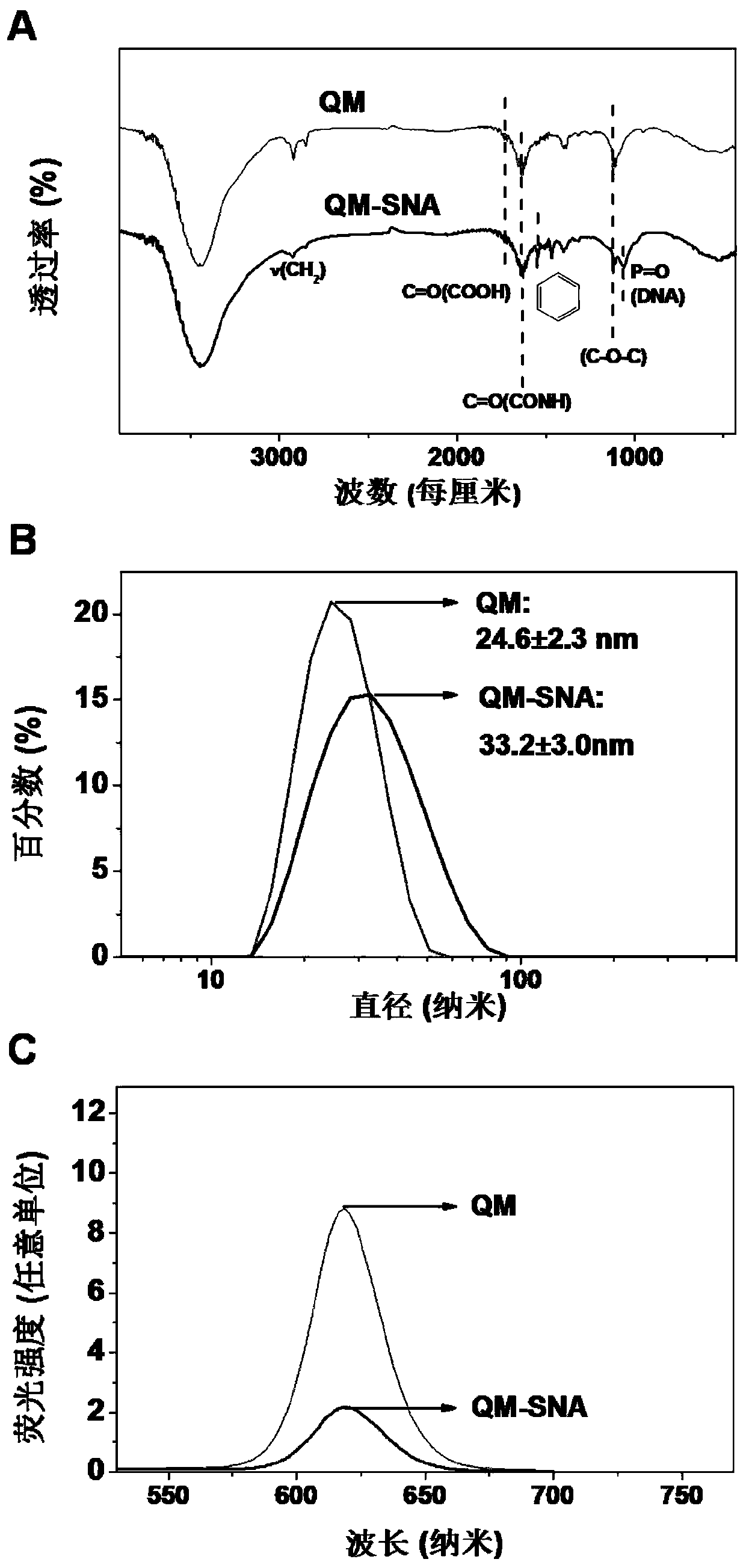
[0031] Characterize the composition, hydrodynamic size, fluorescence and other properties of QM-SNA, the results are as follows figure 2 shown. figure 2 A, Compared with QM, the infrared spectrum of QM-SNA appeared the characteristic peak of CONH bond attributed to the surface coupling (1635cm -1 ), attributed to the P=O bond characteristic peak of nucleotide composition (1062cm -1...
Embodiment 3
[0033] QM surface target-triggered DNase walking
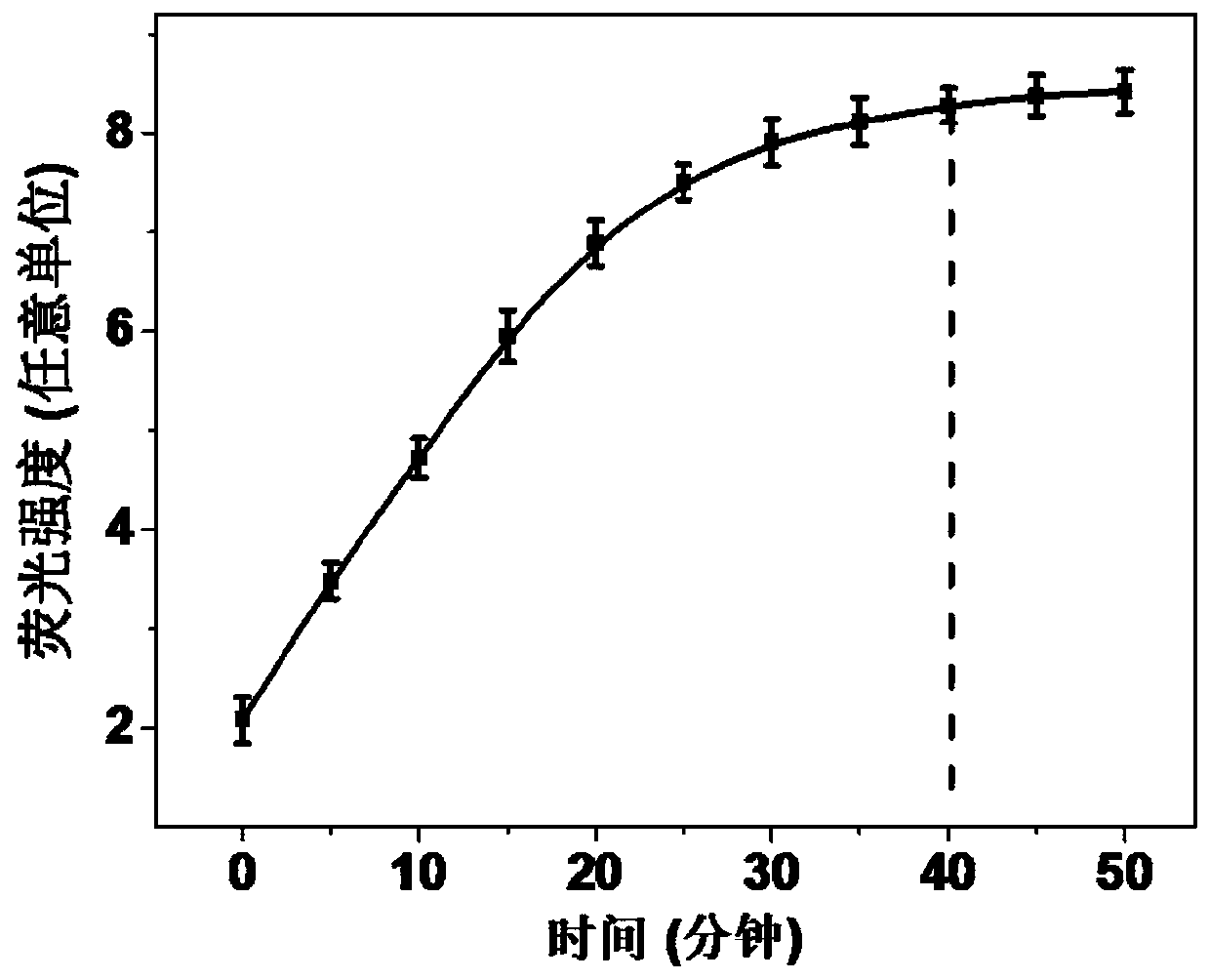
[0034] Add QM-SNA (10nM) and target miRNA (50pM) solution to Tris-HCl (25mM, pH 7.4) buffer solution sequentially, with a total volume of 195μL, and fully react at 37°C for 1h to fully hybridize the target miRNA. Then add 5 μL of Zn 2+ (200mM) solution, measure the fluorescence spectrum every 5min.
[0035] The fluorescence intensity of QM changes as image 3 Shown, Zn 2+ With the extension of time after the addition, the fluorescence of QM gradually increased, and the fluorescence signal reached the maximum at about 40 min, which indicated the continuous walking process of DNase on the surface of QM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com