Method for identifying pathogenic bacteria carrying toxic factors in water body based on metagenome technology
A metagenomic and pathogenic bacteria technology, applied in the field of water ecological security, can solve the problems of limited amplification target gene primer specificity, long culture cycle, difficult pathogenic bacteria, etc., and achieve high reliability results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Sample collection and metagenomic sequencing used in the present invention
[0049] 1.1 Sample collection
[0050] The receiving rivers of two sewage treatment plants in Beijing, Qinghe River and Tonghui River, were selected as the research objects. The basic information of the two sewage treatment plants is shown in Table 1. In December 2017, March, June, and September 2018, the planktonic microorganisms in the water and the biofilm growing on the stones were sampled throughout the year. Set up 7 sampling points for each river, one of which is located at the outlet of the sewage plant, three points are located upstream of the outlet of the sewage plant, and three points are located downstream of the sewage outlet, and the distance between two adjacent sampling points is 200±20 meters . Due to the freezing of the Qinghe River in December 2017, no river water samples were collected. A total of 105 samples were collected, of which 49 were river water samples a...
Embodiment 2
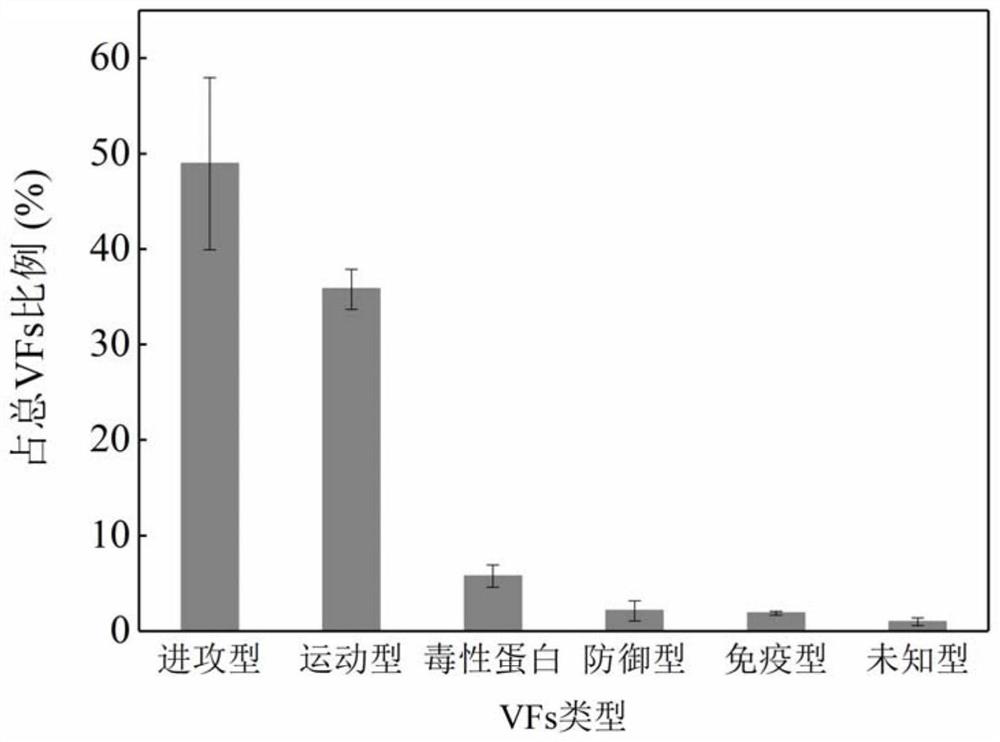
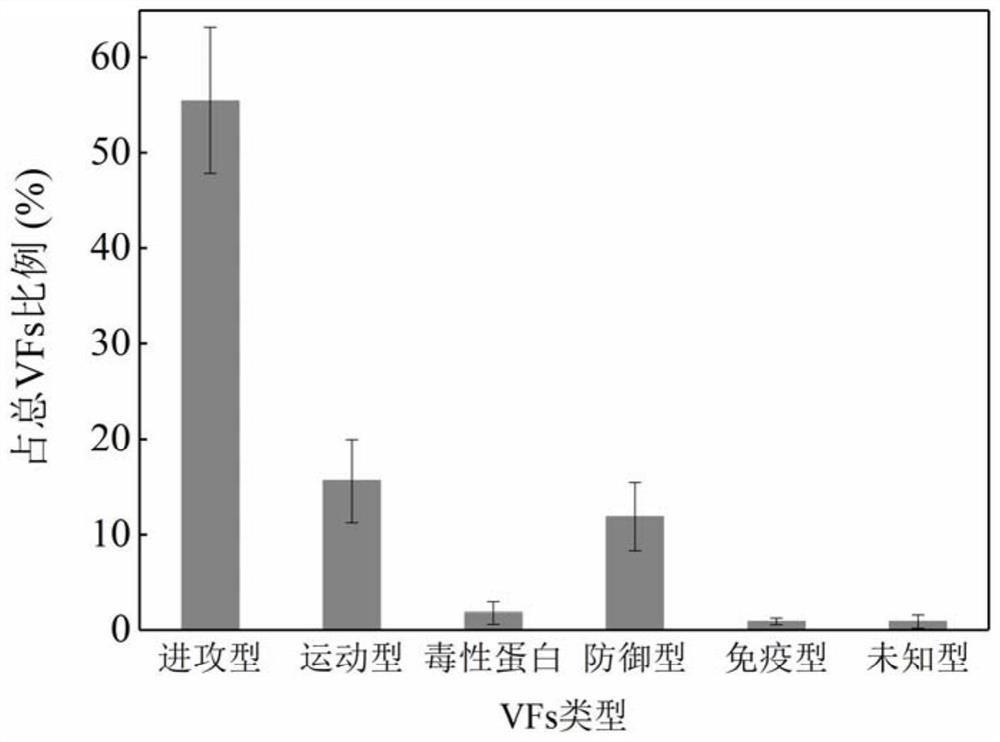
[0059] Embodiment 2 identifies pathogenic bacteria containing virulence factors
[0060] 2.1 Microbial community structure analysis
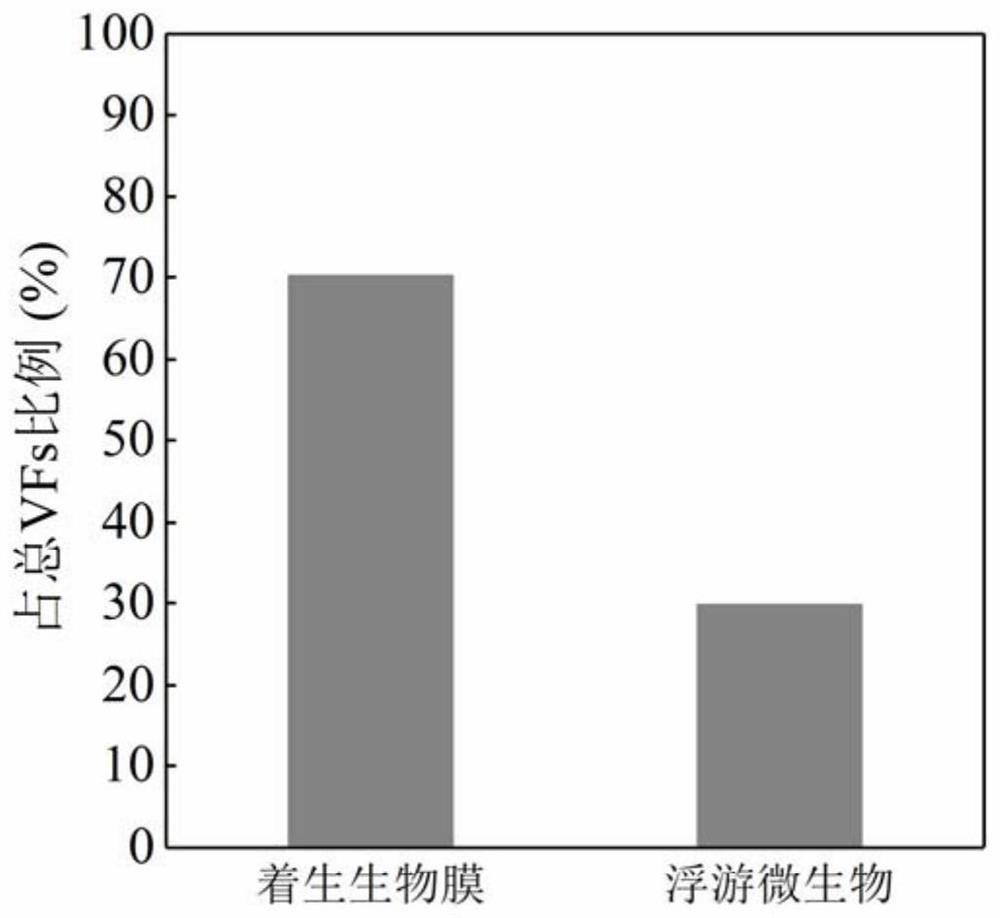
[0061] As described in Example 1, the diversity between the planktonic microbial community and the attached biofilm samples of Qinghe River and Tonghui River was analyzed. The weighted Unifrac distance results are shown in Table 2. The results show that the difference between the planktonic microbial community and the attached biofilm samples is greater than the difference between the two rivers. The follow-up analysis focused on comparing the differences between planktonic microorganisms and attached biofilms.
[0062] Table 2 Analysis results of diversity among samples
[0063]
[0065] Using MEGAHIT (v1.1.3) software to splice and assemble the original metagenomic data, combined with the three binning methods of MetaBAT 2, MaxBin 2.0 and CONCOCT, and applying the MetaWRAP binning process, based on completene...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com