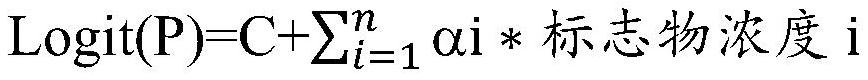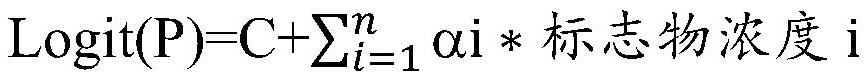Method for constructing mathematical model for detecting breast cancer in vitro, and application thereof
A mathematical model, breast cancer technology, applied in the field of medical diagnosis, to achieve the effect of improving precision and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Using the self-made chemiluminescence detection kit or the self-made flow cytometry fluorescence luminescence detection kit, the concentrations of 11 breast cancer protein markers (CA15-3, TIMP-1, PAI, NMP66, OPN, CEACAM6, RARA, IGFBP1, GRM1, GRIK1, S100A8), the concentration of 13 breast cancer molecular markers (UHRF1, BRCA1, FGFR2, HER-2 / neu, STAT3, Ki-67, PIK3CA, CCNB1 (Cyclin B1), MYBL2, ACTB (b-actin), CTSL2 (Cathepsin L2), CD68, BAG1), the concentrations of 12 breast cancer-related DNA methylation markers (PITX2P2, GSTP1, RASSF1A, RAR-β2, DNMT1, DMAP1, MeCP2, MBD1, CDKN2B, CDKN2A, p21WAF1 / CIP1, RARβ2).
[0031] Perform logistic regression analysis on the test concentrations of the above-mentioned relevant markers to obtain Logit(P)=constant+λ1*P1+λ2*P2+η3*P3+η4*P4...
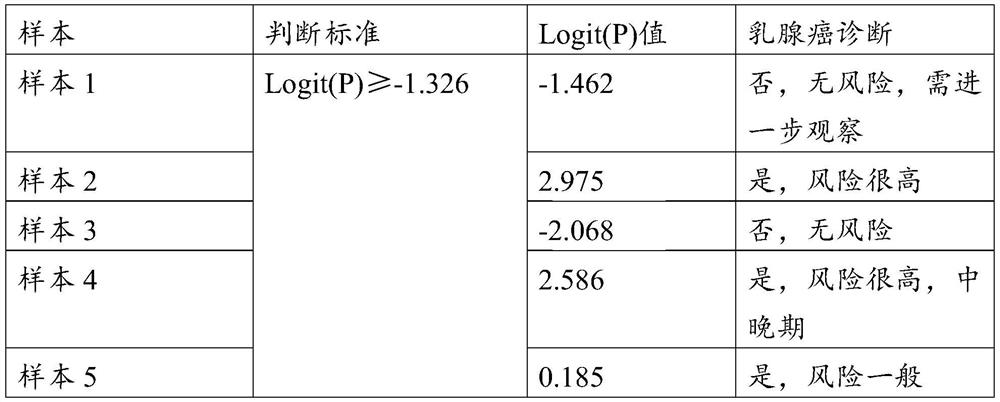
[0032] Then test the concentration of each marker in the unknown blood sample and substitute it into the regression model. According to the calculated Logit (P) and the judgment standard of the re...
Embodiment 2
[0035] Use the purchased or self-made chemiluminescence method kit to test the concentration of 6 breast cancer protein markers (CA15-3, TIMP-1, OPN, CEACAM6, CEA, IGFBP1) in the blood sample, and use the fluorescence in situ hybridization method to test the concentration of the six breast cancer protein markers in the blood sample. 9 molecular markers of breast cancer (miR-21, miR-20a, miR-214, miR-181a, miR-1304, miR-141, miR-200a / c, miR-203, miR-210), with purchased The concentration of 13 breast cancer autoantibodies (CTAG1B, CTAG2, TP53, RNF216, PPHLN1, PIP4K2C, ZBTB16, TAS2R8, WBP2NL, DOK2, PSRC1, MN1, TRIM21) in blood samples was detected by immunofluorescence method, and urine by flow cytometry method or 11 breast cancer-associated exosomes (miR-27a, miR-451, miR-21-5p, miR-21, miR-221, TGF-β1, HMGB1, CagA, GKN1, UBR2, TRIM3) in blood, using Fluorescence flow method to detect 7 related inflammatory factors and growth factors in urine or blood: (CRP, Ch17CEP, sHER2, MAD...
Embodiment 3
[0040] Breast cancer protein markers are CA15-3, TIMP-1, uPA, PAI, NMP66, OPN, CEACAM6, Bc1, IGFBP1, breast cancer molecular diagnostic markers are UHRF1, BRCA1, BRCA2, FGFR2, STAT3, PIK3CA, MYBL2, GAPDH , RPLPO, BCL2, breast cancer-related DNA methylation markers are PITX2P2, APC, GSTP1, RASSF1A, RAR-β2, DNMT1, obtain the concentration values of these markers in the sample, perform natural logarithmic transformation, and eliminate After no contributing markers, the resulting regression model is: Logit(P)=-2.43+1.012*Ln(CA15-3)+0.452*Ln(uPA)+0.785*Ln(OPN)+0.652*Ln(CEACAM6) +0.741*Ln(IGFBP1)+1.210*Ln(UHRF1)+0.471*Ln(BRCA1)+0.723*Ln(FGFR2)+0.457*Ln(PIK3CA)+0.789*Ln(RASSF1A)+0.354*Ln(PITX2P2)+ 0.987*Ln(APC)+0.541*Ln(GSTP1), where Ln is the natural logarithm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com