Nasopharyngeal carcinoma microRNA detection kit
A technology of hsa-mir-34b-3p and hsa-mir-34c-5p, which is applied in the field of molecular biology, can solve the problems of low specificity, weak repeatability and poor stability, and achieves high specificity and good repeatability. , good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] This example provides a method for preparing a capture probe. The base sequence of the capture probe designed in this example is, from the 5' end to the 3' end, the specific sequence P1 that binds to the target microRNA to be detected, the spacer arm sequence, P2 sequence.
[0021] The spacer sequence can separate the P2 sequence of the capture probe from the target microRNA to be detected. By setting the spacer sequence of an appropriate length inside the probe, it can reduce steric hindrance, improve the efficiency and specificity of the hybridization reaction sex. The spacer sequence of the capture probe of the present invention is preferably 5 Ts.
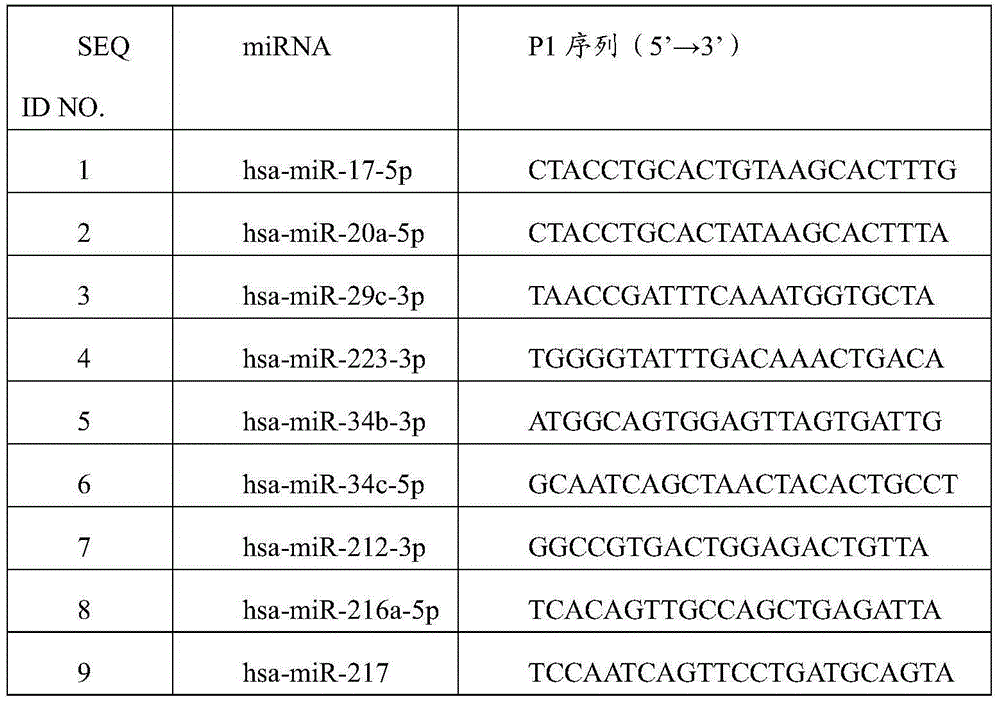
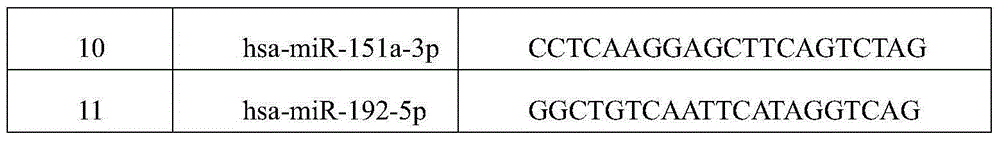
[0022] This example is aimed at hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR-29c-3p, hsa-miR-223-3p, hsa-miR-34b-3p, hsa-miR-34c- 5p, hsa-miR-212-3p, hsa-miR-216a-5p, hsa-miR-217, hsa-miR-151a-3p and hsa-miR-192-5p designed capture probes, see Table 1, Table 1 2:
[0023] Table 1 P1 sequence of capture probe
[0024]
...
Embodiment 2
[0029] This embodiment provides the preparation method of the signal amplifying component, the signal amplifying component obtained by the preparation method described in this embodiment is selected from: the first signal amplifying component, the second signal amplifying component and the third signal amplifying component Any one; the first signal amplification component includes a primary signal amplification probe, and the 3' end of the first signal amplification component is also modified with a fluorescent group; the second signal amplification component is a primary signal amplification probe and secondary signal amplification probe, the 3' end of the second signal amplification component is also modified with a fluorescent group; the third signal amplification component is a primary signal amplification probe, a secondary signal amplification In the amplification probe and the tertiary signal amplification probe, the 3' end of the third signal amplification component is ...
Embodiment 3
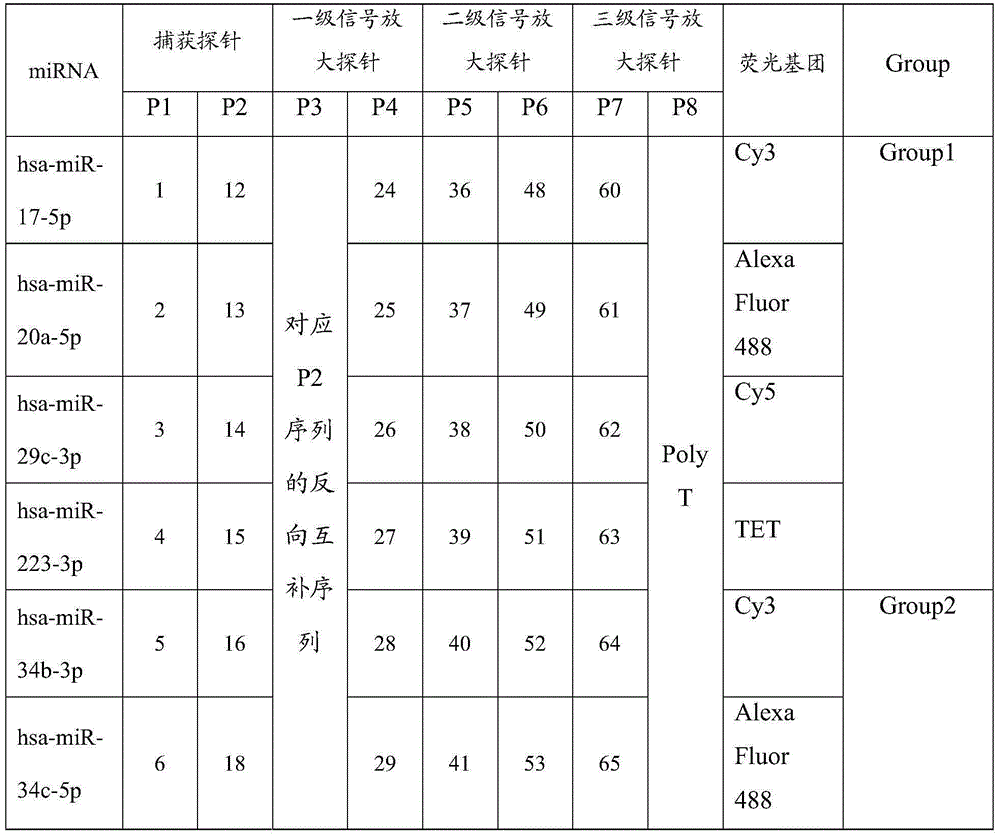
[0056] This embodiment provides a preparation method of nasopharyngeal carcinoma microRNA detection kit, said nasopharyngeal carcinoma microRNA detection kit comprises: the probe component prepared in Example 1 and the signal amplification component prepared in Example 2 .
[0057] The composition of a nasopharyngeal carcinoma microRNA detection kit designed in this embodiment is shown in Table 7.
[0058] Table 7 A nasopharyngeal carcinoma microRNA detection kit (table number is SEQ ID NO.)
[0059]
[0060]
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com