A self-cleavage resistant and high specific activity trypsin mutant
A technology of trypsin and trypsinogen, applied in the directions of enzymes, peptidases, hydrolase, etc., can solve the problems of affecting the application efficiency and level, and the decline of enzyme activity, and achieves the improvement of easy self-cleavage and reduced activity, high specific activity, good The effect of anti-self-cutting properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1. Cloning of porcine trypsinogen gene.
[0058] According to the amino acid sequence of wild porcine trypsinogen, Pichia pastoris was selected as the expression host for codon optimization to obtain the optimized gene of Pichia trypsinogen (SEQ ID NO. 1), and the wild-type trypsinogen gene (SEQ ID NO. 1) was synthesized. .1), design primers and amplify the gene by PCR technology, use T4 ligase to connect the amplified gene fragment to the vector pMD19T to obtain the recombinant cloning vector pMD19T-try. Transform into E. coli JM109 competent cells.
Embodiment 2
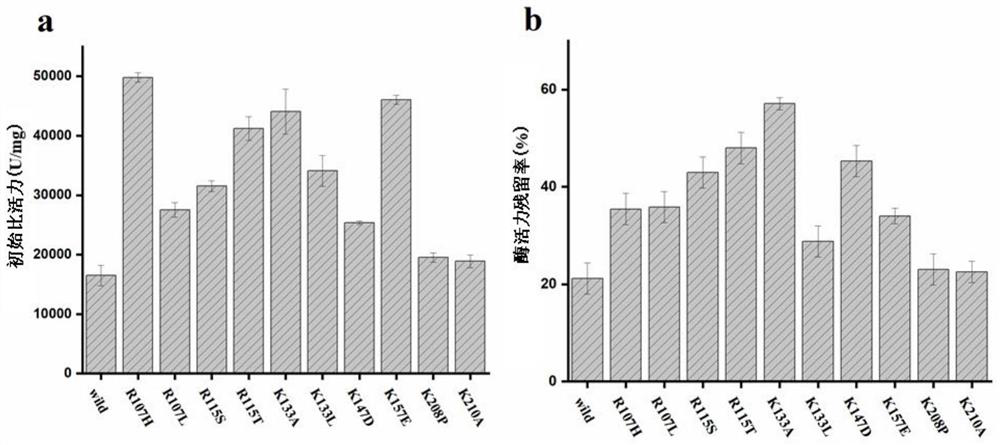
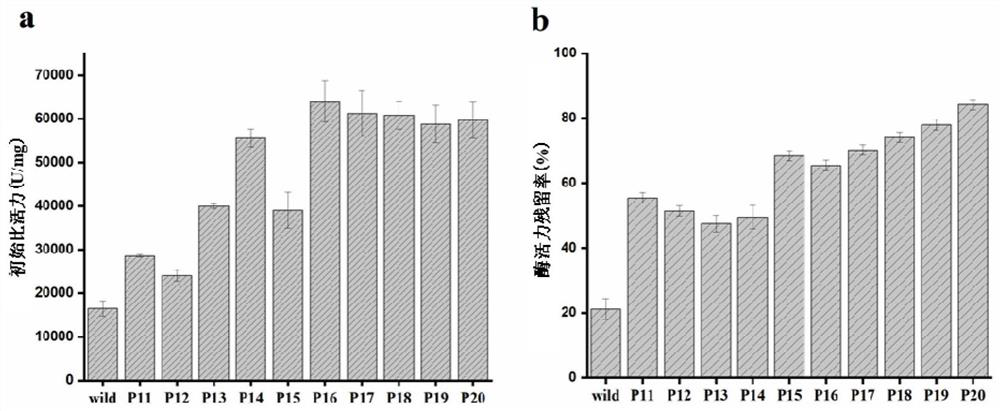
[0059] Example 2. Site-directed mutagenesis
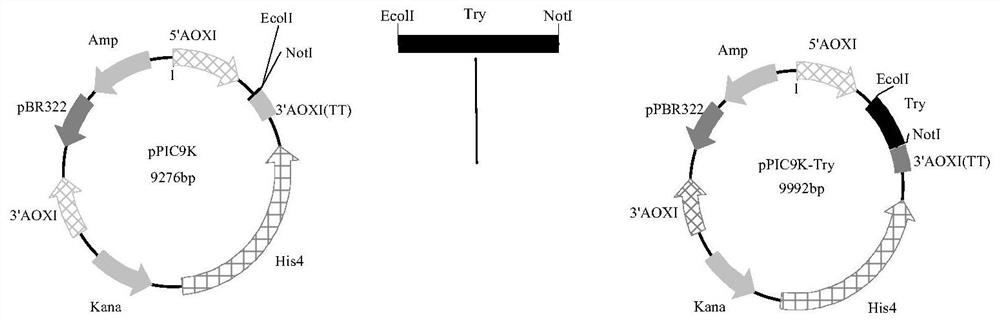
[0060] The process of site-directed mutagenesis is as follows: using the constructed PMD19T-try as a template, design corresponding primers for inverse PCR, at the corresponding amino acid positions 107, 115, 133, 147, 157, 208 and 210. Single point or combined mutation was introduced, and the primer sequences are shown in Table 2. Use Dpn I restriction endonuclease to remove the original template, carry out agarose electrophoresis to verify the amplified product and purify the recovered product, transform the recovered plasmid into E. coli, extract the plasmid from the constructed strain, and perform single enzyme digestion and double-enzyme digestion to verify and sequence, and extract the recombinant plasmid pMD19T-M-try (M represents the corresponding mutant); the constructed recombinant plasmid pMD19T-M-try (M represents the corresponding mutant) and the expression vector pPIC9K were respectively used Ecol I and Not I were do...
Embodiment 3
[0064] Embodiment 3, recombinant Pichia pastoris shake flask fermentation
[0065] The strains were inoculated into YPD solid medium for three-district streaking, and cultured at 28°C for 2 days; a single colony was picked from the YPD solid plate to YPD liquid medium, 28°C, 200rpm for 24h; YPD liquid culture was inoculated at 2% of the inoculum The well-cultivated seed liquid was cultured to BMGY medium, 28°C, 200rpm to the bacterial liquid OD 600 For 2-6, then transfer it to a sterile centrifuge tube and centrifuge at 5000rpm for 10min to retain the bacteria; add an appropriate amount of BMMY and repeat the centrifugation to remove residual glycerol; transfer to BMMY medium, add methanol to a final concentration of 0.5%; then every 12h Add methanol with a final concentration of 0.5% to induce culture for 96 h.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com