Multiplex PCR primer group and next-generation sequencing database building method for MLST (multilocus sequence typing) tracing of vibrio parahaemolyticus
A hemolytic vibrio and next-generation sequencing technology, applied in the field of microbial detection, can solve problems such as heavy workload and complicated operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
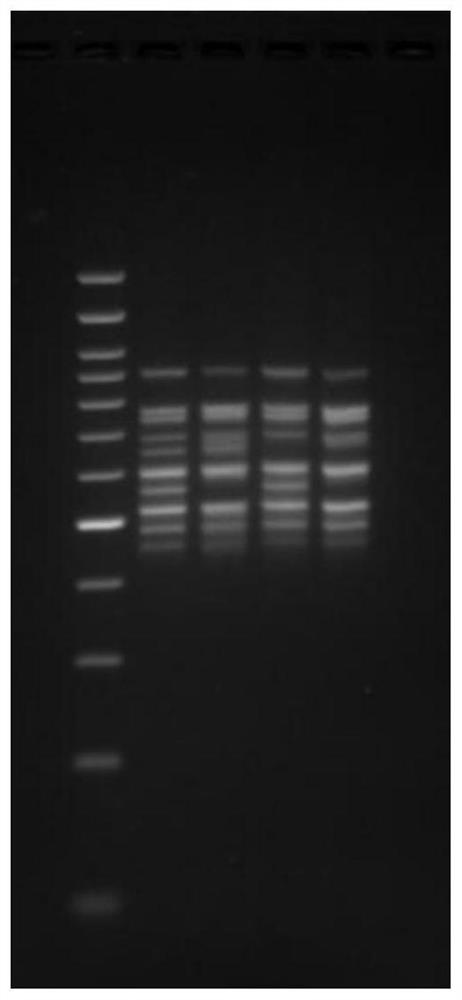
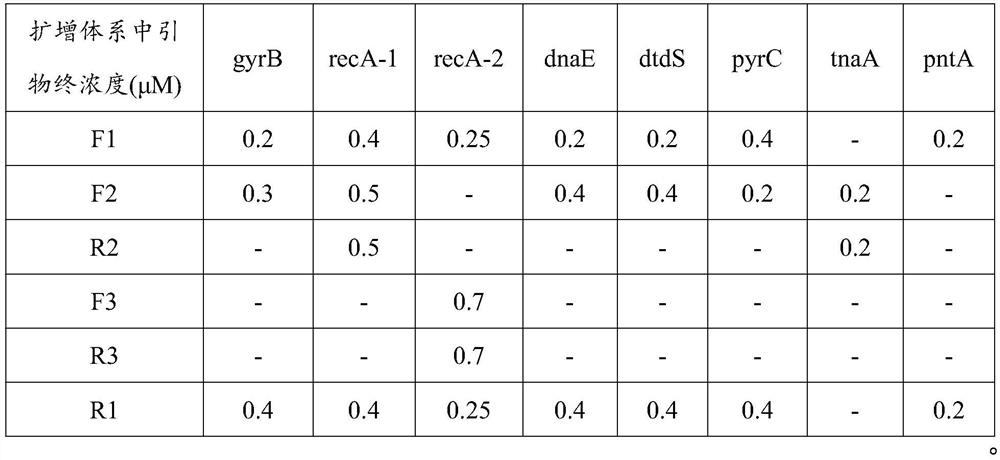
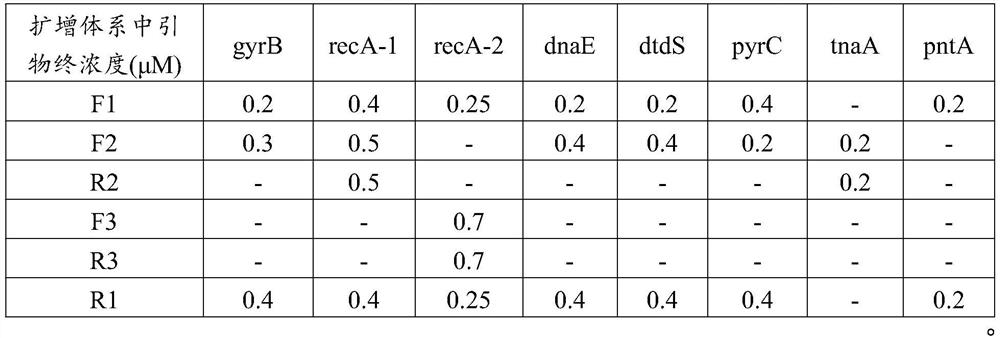
[0056] 1. The basic information of the seven housekeeping genes and the primers for library construction are shown in Table 1.
[0057] 2. Database construction plan
[0058] 2.1 One round of primer synthesis
[0059] In addition to the sequence used for multiplex PCR amplification, the one-round primer will also have a general sequence for the second round of primer-binding amplification. Take the tnaA gene primer as an example:
[0060] F1: ACACTCTTTCCCTACACGACGCTCTTCCGATCT CGGTATCGATGGAAAACATGC (SEQ ID NO. 23);
[0061] R1: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC GCAATATTTTTCGCCGCATCAAC (SEQ ID NO. 24);
[0062] Among them, ACACTCTTTCCCTACACGACGCTCTTCCGATCT (SEQ ID NO.25) is the universal sequence of the F-terminal primer: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC (SEQ ID NO.26) is the general sequence of the R-terminal primer.
[0063] 2.2 DNA extraction method of the sample to be tested
[0064] The Vibrio parahaemolyticus isolates obtained from the port samples, numbered 1...
Embodiment 2
[0097] The four sets of libraries prepared in Example 1 were mixed and then sequenced, see Table 9 for details.
[0098] Table 9 High-throughput sequencing library information (equal-quality mixed sample sequencing of 4 groups of libraries)
[0099]
[0100] Analysis of sequencing results
[0101] 1. See Table 10 for data quality.
[0102] Table 10 Summary of library sequencing data quality prepared by different methods
[0103]
[0104]
[0105] 2. See Table 11 for gene sequence matching.
[0106] Table 11 Matching of library gene sequences prepared by different methods
[0107]
[0108] 3. Analysis of sequencing results
[0109] 1) 1269 strain, two sets of amplification schemes, all seven gene sequences were detected, all of which could be matched to the corresponding gene numbers, and the comparison results of the two sets of data were consistent.
[0110] 2) For the 1177 strain, two sets of amplification schemes, seven gene sequences were detected, all of wh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com