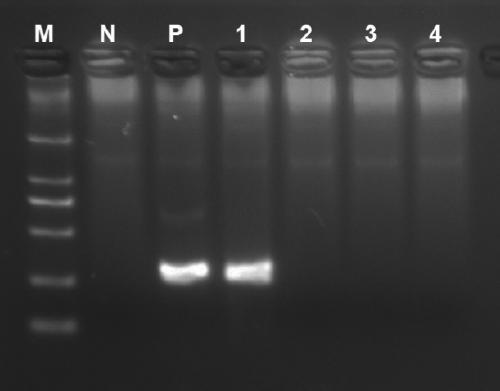
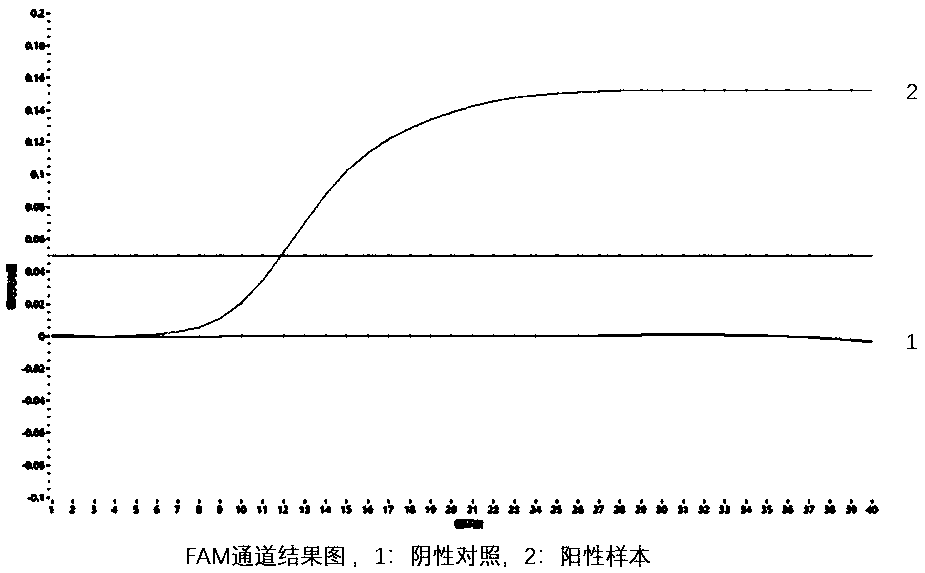
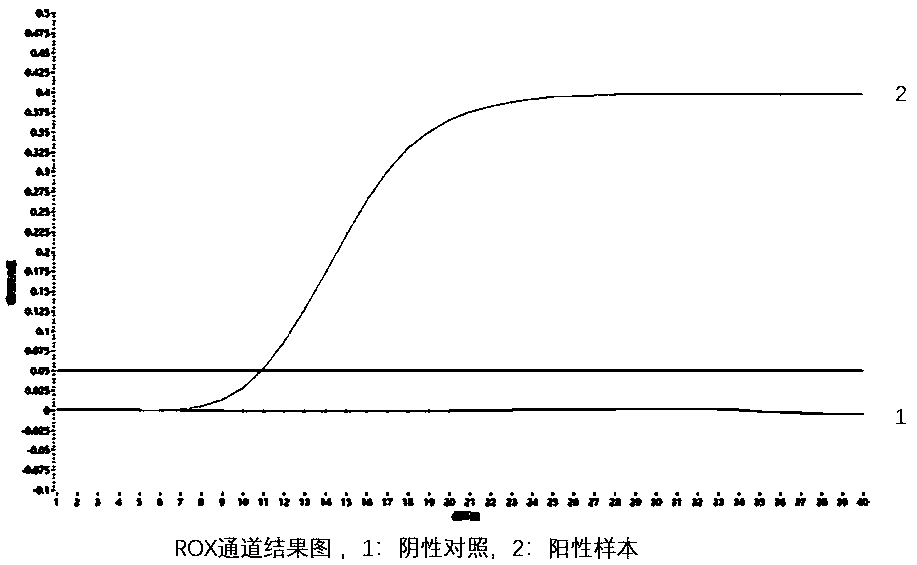
RDA (Recombinase-dependent amplification) method and kit for quickly detecting Coxsachie virus 16 and enterovirus 71
A technology for coxsackie virus and enterovirus, applied in the field of molecular biology, can solve the problems such as the inability to meet the simple, rapid and accurate detection requirements of disease prevention and control, the long cycle of virus isolation and detection methods, and the high operational requirements of detection personnel. , to achieve the effect of low detection cost, high precision and rapid molecular detection, and low length requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1 A detection kit for coxsackievirus A16 (CA16) and enterovirus 71 (EV71) RDA fluorescence method
[0093] (1) Acquisition of recombinant enzyme KX and KY proteins
[0094] The reported recombinant enzyme UvsX has poor stability and is difficult to mass produce and store for a long time. In order to solve this problem, the research and development team used bioinformatics methods to analyze and simulate large quantities of protein structures, and finally found a new recombinant enzyme Enzyme KX and its accessory protein KY.
[0095] In this example, the R&D team extracted the key functional site information in the recombinase structure, such as DNA binding site, ATP hydrolysis site, etc., and mapped it to the three-dimensional structure of the protein to obtain secondary structure information and tertiary structure information , by integrating the functional residues of the primary structure sequence, secondary structure features and tertiary structure spatial ...
Embodiment 2
[0145] Example 2 RDA fluorescence method detection reagent sensitivity test
[0146] The positive control is the pUC57 plasmid containing the conserved genes of coxsackievirus type A16 and enterovirus type 71 (EV71), and the negative control is the empty vector pUC57 plasmid.
[0147] The specific operation is as follows:
[0148] Step 1: Dilute the positive control plasmid to 10^4c, and then dilute to 10^3c, 10^2c, and 10^1c by 10-fold serial dilution.
[0149] Step two, sample processing. Take 5 μL of each concentration of the plasmid in step 1 in an EP tube, and take 5 μL of the negative control in another EP tube, add 20 μL of Buffer A respectively, shake and mix, and let stand at room temperature for 10-15 minutes;
[0150] Step 3, system preparation and testing. Add 25 μL Buffer B to each tube, shake and mix, add 50 μL of the mixed solution to the RDA fluorescence method reaction module, cover the tube cap, shake and centrifuge, and detect immediately; the reaction pr...
Embodiment 3
[0162] Example 3 RDA fluorescence method detection reagent specificity test
[0163] 1 case of Coxsackie virus 16 (Coxsachie virus 16, CA16), 1 case of enterovirus 71 (Enterovirus, EV71), 1 case of Norovirus (NV), 1 case of adenovirus (Adenovirus, AV), rotavirus (Rotavirus, RV) 5 kinds, a total of 5 cases were tested by fluorescent quantitative PCR as positive samples for the corresponding virus, and the specificity of the kit was tested.
[0164] The specific operation is as follows:
[0165] Step 1. Sample processing. Take 5 μL of each of the above 5 positive samples in EP tubes, and at the same time take 5 μL of the positive control and negative control of the kit in a new EP tube, add 20 μL Buffer A respectively, shake and mix, and let stand at room temperature for 10-15 minutes;
[0166]Step 3, system preparation and testing. Add 25 μL Buffer B to each tube, shake and mix, add 50 μL of the mixed solution to the RDA fluorescence method reaction module, cover the tube ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com