Method for screening and identifying stress response gene expression regulatory factors
A technology for regulating factors and gene expression, applied in the biological field, can solve the problems of identification and determination that have not been developed, and achieve the effect of reducing time and labor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
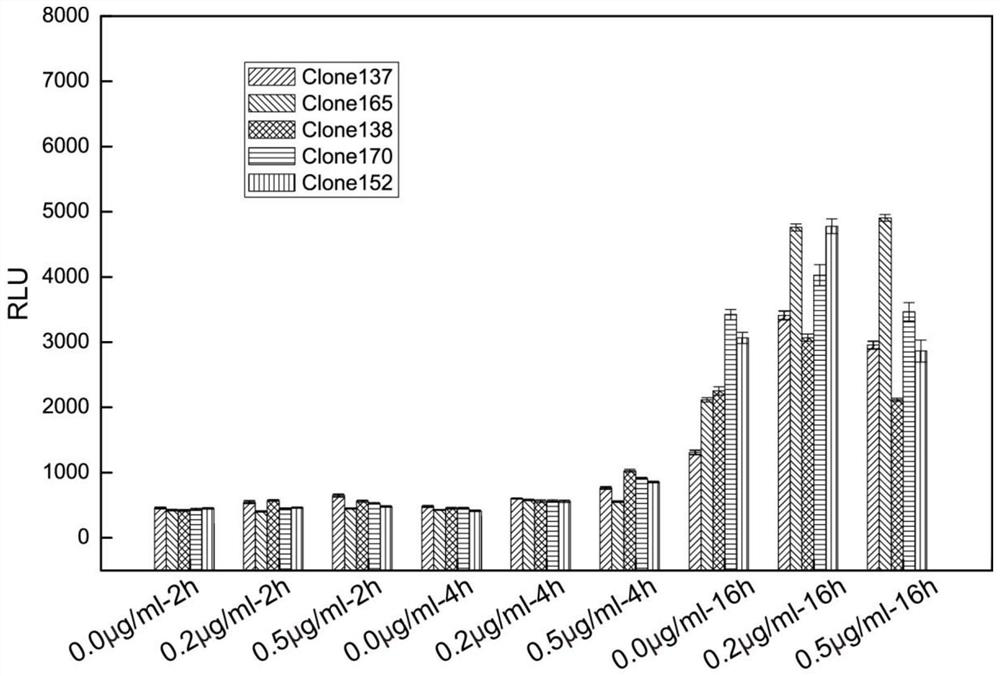
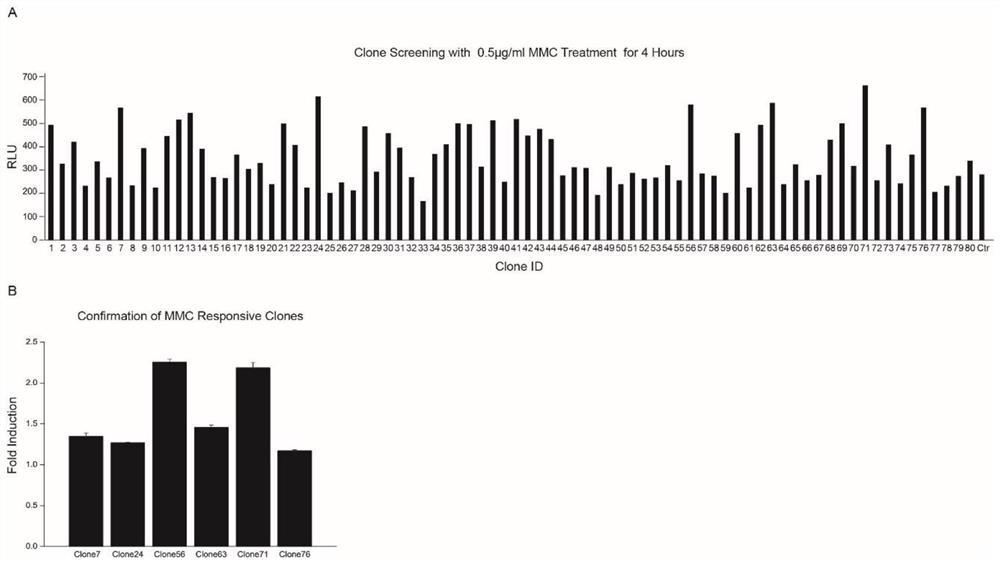
Embodiment 1
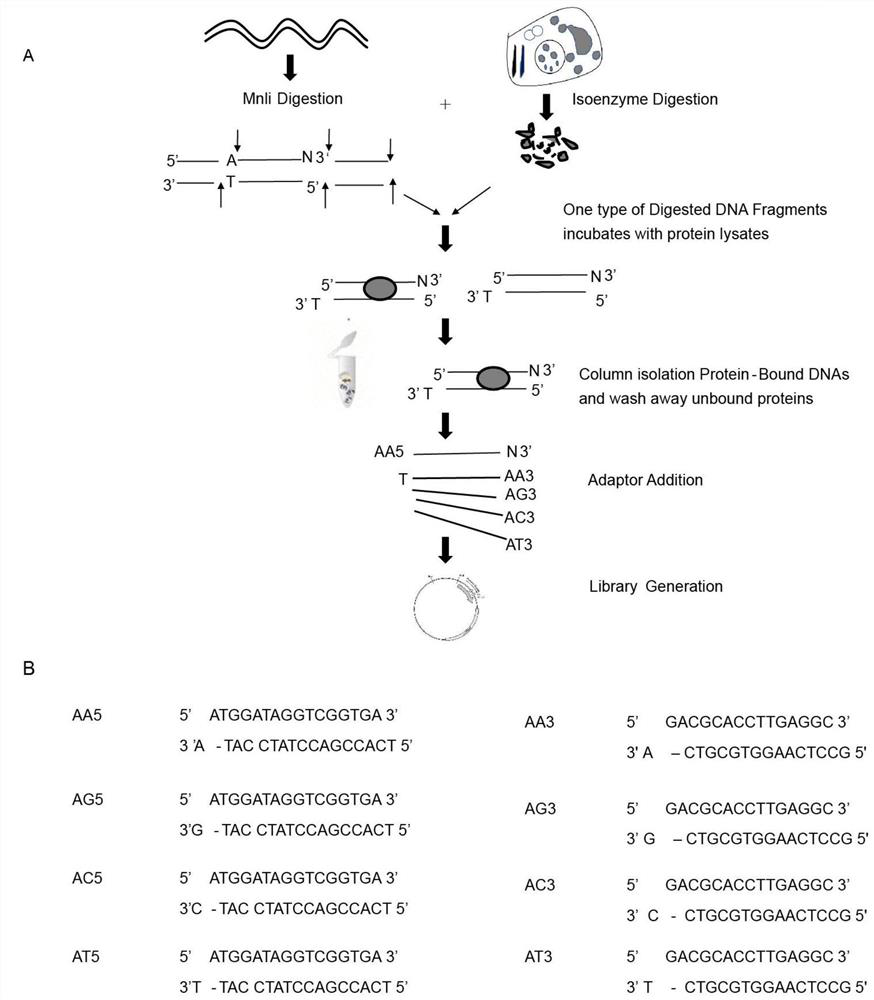
[0040] 1. Materials and methods
[0041] 1. Preparation of cell lysate
[0042] 1 mL of DH5α culture was centrifuged at 10,000 g for 1 min, and the pellet was resuspended in 300 μL of lysis buffer (10 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) and 0.1% (w / v ) Polyethylene glycol octylphenyl ether (Triton X-100)). 7.5 μL of freshly made lysozyme solution (10 mg / mL in 10 mM Tris-HCl, pH 8.0, final concentration = 0.25 mg / mL) was added and mixed by tapping the tube, and the lysis mixture was incubated at room temperature for 10-20 minutes. After centrifugation, the supernatant was used for affinity screening.
[0043] 2. Affinity screening
[0044] DH5α cells were collected by centrifugation, resuspended in 200 μL lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5% SDS), and treated with 20 μg / mL proteinase K at 55° C. for 2 h. Genomic DNA was extracted with phenol and chloroform. Genomic DNA was digested with Mnli,5'...CCTC(N)7...3', whic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com