Method for constructing tumor mutation load TMB panel and using method of tumor mutation load TMB panel
A mutation load and panel technology is applied in biochemical equipment and methods, medical data mining, and microbial determination/inspection. The effect of low sequencing cost, short turnaround time, and long detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
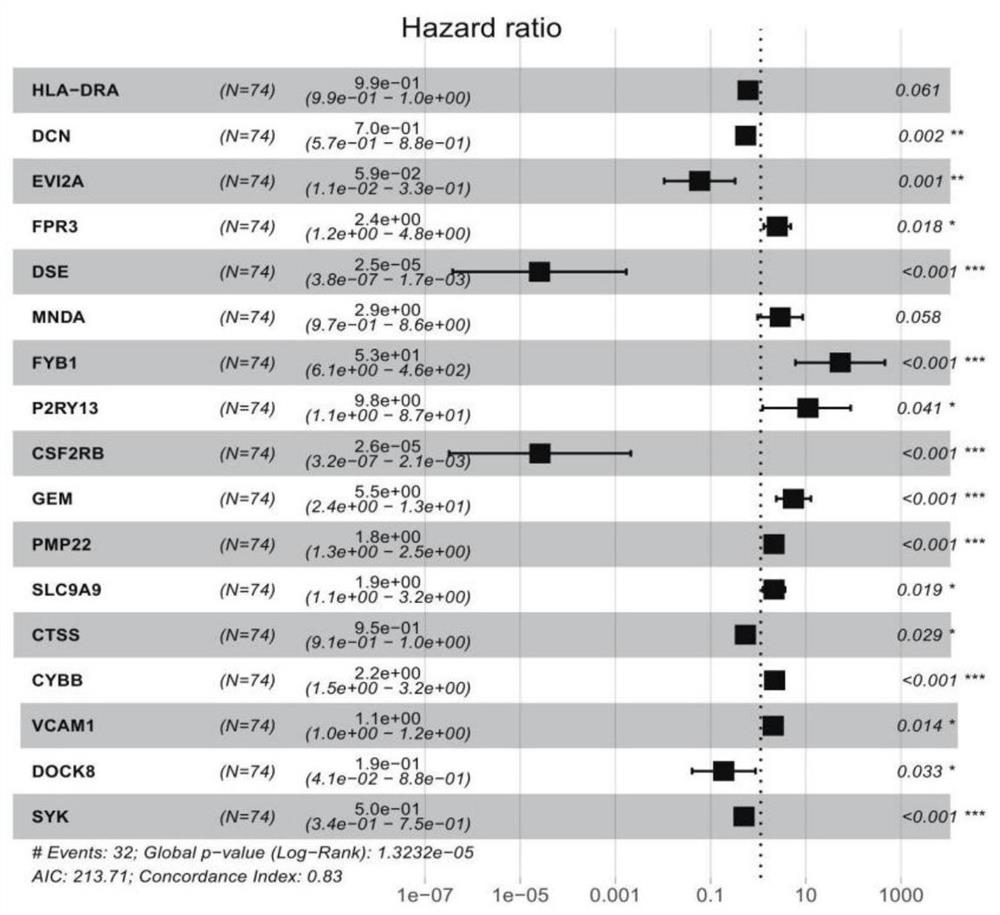
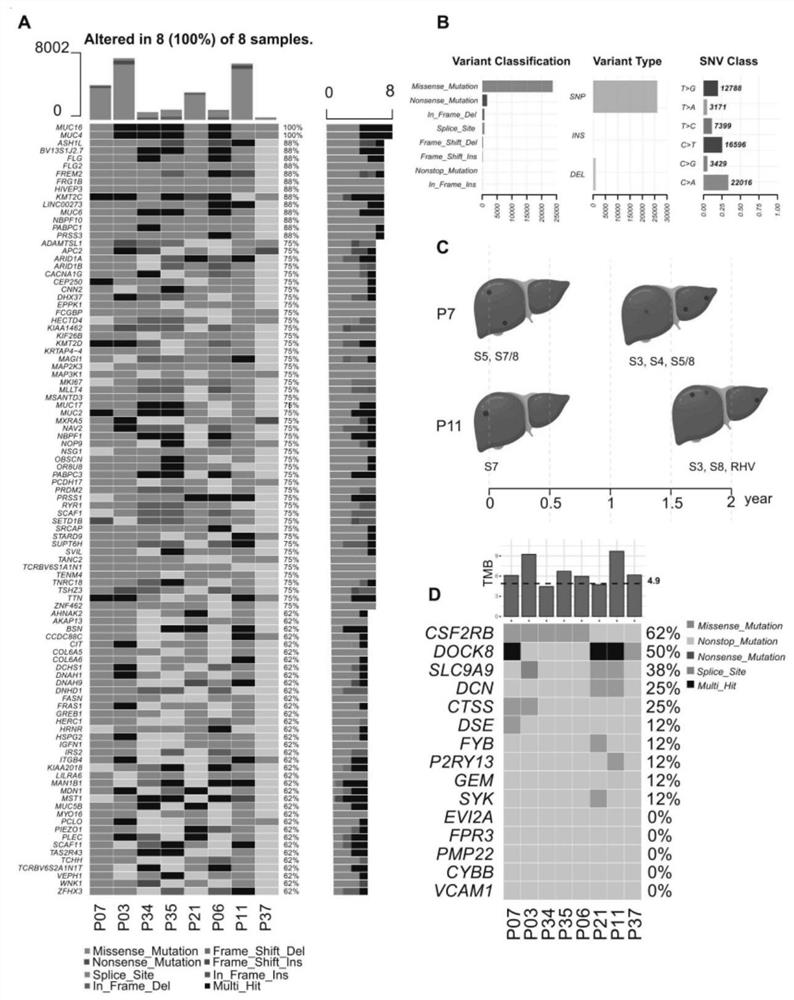
[0023] The method for constructing a tumor mutation load TMB panel of the present invention comprises the following steps:
[0024] 1) Acquire the transcriptome data of tumor patients, the transcriptome refers to the quantitative transcriptome of miRNA expression, and divide the transcriptome data into high tumor mutation burden group and low tumor mutation burden group according to the optimal tumor mutation burden threshold affecting patient survival mutation load group;
[0025] 2) Screen the immune-related differentially expressed gene DEGs between the high tumor mutation burden group and the low tumor mutation burden group in step 1), and perform enrichment analysis on the gene ontology of the DEGs to obtain the protein interaction PPI between the differential genes Information, draw a PPI network diagram, and use this to filter out several top-ranked nodes;
[0026] 3) The transcriptome expression data of the high tumor mutation burden group and the low tumor mutation b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com