Method for identifying authenticity of cucurbita pepo variety
A technology of zucchini and locus, which is applied in the field of identifying the authenticity of zucchini varieties and can solve the problems of lack of DNA molecular detection methods and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1. Acquisition of primer combinations for identifying the authenticity of zucchini varieties
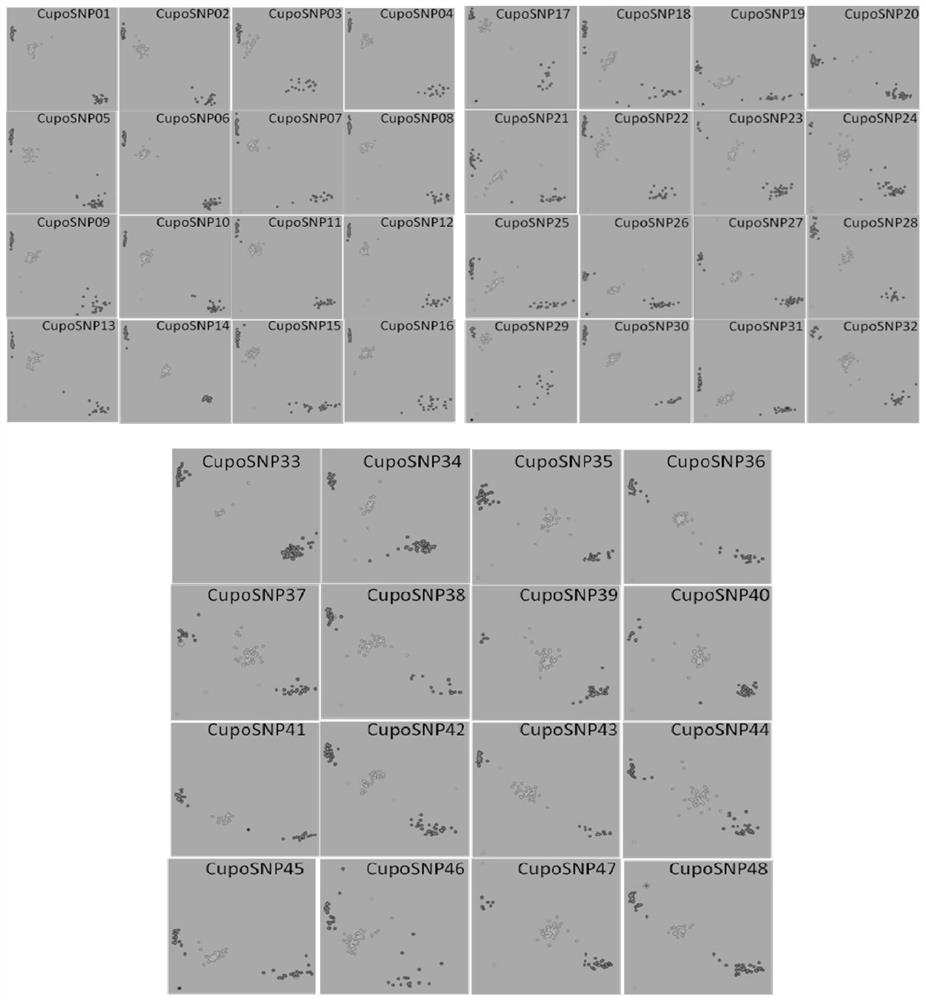
[0039] 1. Discovery of 48 SNP sites
[0040] The present invention obtains 48 SNP sites based on the resequencing data of 30 zucchini representative resources. These 30 zucchini represent rich types of resources, covering mosaic zucchini (8 parts), seedless zucchini (7 parts), green-skinned zucchini (8 parts) and long-vine zucchini (7 parts), basically including the main ecology of zucchini type, with high genetic diversity in terms of agronomic traits, reflecting germplasm representation as much as possible.
[0041] Specifically, the screening criteria for SNP loci are as follows: evenly selected positions throughout the genome, good polymorphism, low heterozygosity, MAF>0.3, good PCA clustering effect, high discrimination, and conservative 50bp sequences on both wings (no InDel , no SSR, no other SNP).
[0042] For the basic information of the 48 SNP sites, see c...
Embodiment 2
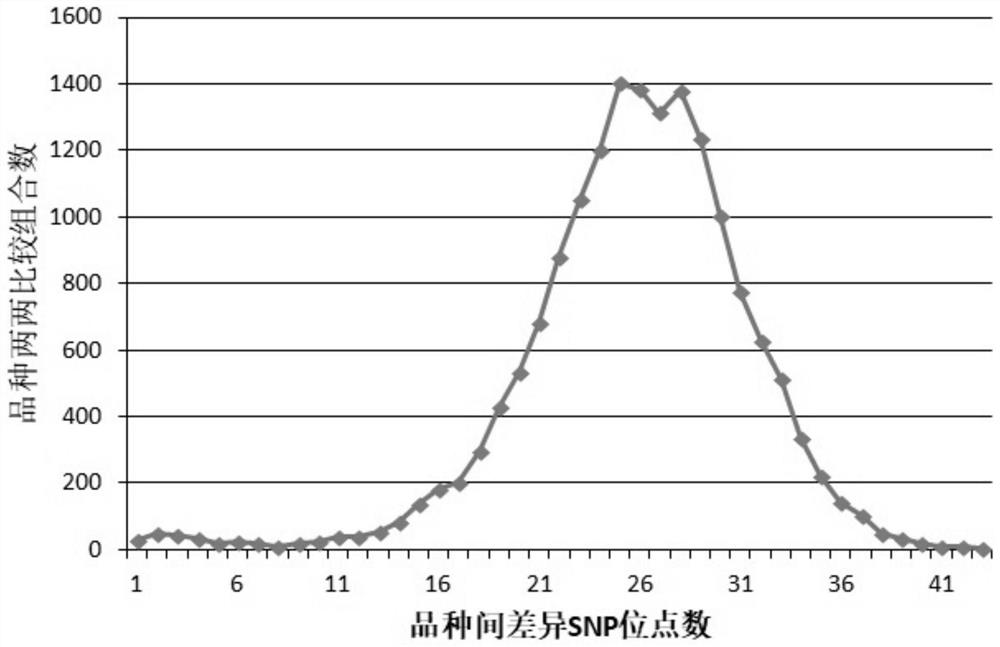
[0057] The validity check of the primer combination that embodiment 2, embodiment 1 develop
[0058] Randomly select 265 zucchini varieties for testing to check the effectiveness of the primer combinations developed in Example 1.
[0059] The basic information of the 265 tested zucchini varieties is shown in Table 3. The 265 tested zucchini varieties are all common fine varieties or some imported varieties.
[0060] Table 3. Names and sources of 265 tested zucchini varieties
[0061]
[0062]
[0063]
[0064]
[0065] 1. Obtaining the genomic DNA of the tested zucchini varieties
[0066] Genomic DNA of 265 leaves (true leaves of 30 seeds mixed) of 265 tested zucchini varieties were extracted by CTAB method to obtain genomic DNA of tested zucchini varieties.
[0067] The quality and concentration of the genomic DNA of the tested zucchini varieties must meet the requirements of PCR, and the standard is: agarose electrophoresis shows a single DNA band without obvio...
Embodiment 3
[0079] Embodiment 3, establishing the method for detecting which variety the zucchini to be tested belongs to among the 265 zucchini varieties in embodiment 2
[0080] One, establish the method for detecting which kind the zucchini to be tested belongs to among the 265 zucchini varieties in embodiment 2
[0081]1. Obtaining the genomic DNA of the zucchini to be tested
[0082] According to the method of step 1 in Example 2, "leaves of the tested zucchini variety" was replaced with "leaves of the zucchini to be tested", and other steps were kept unchanged to obtain the genomic DNA of the zucchini to be tested.
[0083] 2. Using the genomic DNA of the zucchini to be tested as a template, 48 primer sets were used for PCR amplification to obtain PCR amplification products. In each PCR reaction system, the concentration ratio of the primers containing "F1" in the name, the primers containing "F2" in the name and the primer containing "R" in the name is 2:2:5.
[0084] The reactio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com