Kit for detecting nuclear factor-kappa B and application of kit
A nuclear factor and kit technology, applied in the field of nuclear factor-κB detection kits, can solve the problems of low sensitivity, complicated design, and long time consumption, and achieve high sensitivity, low detection limit, and overcome interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
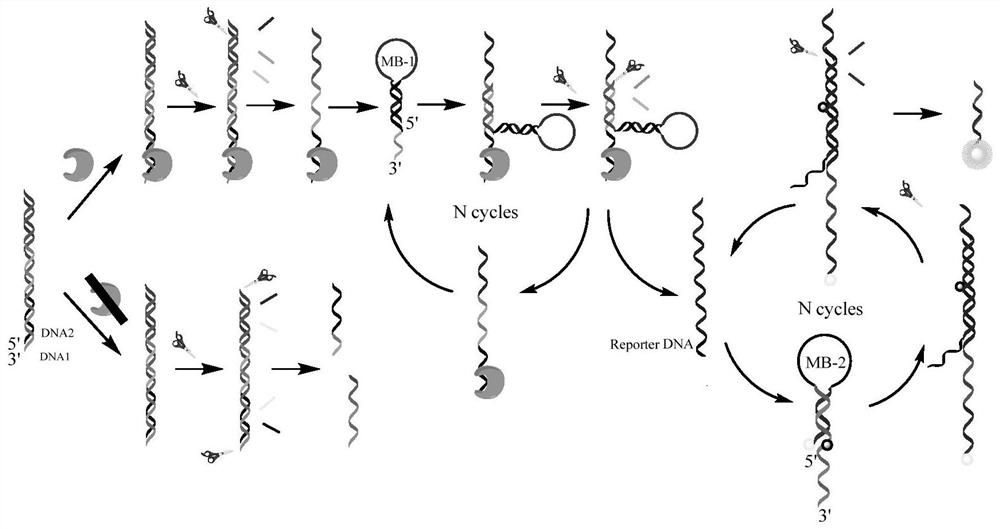
[0042] Mix DNA-1 (SEQ ID No.1) dissolved in DNA reaction buffer and DNA-2 (SEQ ID No.2) dissolved in DNA reaction buffer to obtain a double-stranded probe stock solution with a concentration of 5 μM . Prepared by dissolving molecular beacons MB-1 (SEQ ID No.3) and MB-2 (the 5' end of SEQ ID No.4 is marked with Cy3, and the 20th position is marked with BHQ-2) in DNA reaction buffer Both MB-1 and MB-2 concentrations were 5 μM stock solution. All solutions were heated at 95°C for 5 minutes, then slowly cooled to room temperature and allowed to stand for at least 4 hours.
[0043] When detecting NF-κB p65, add NF-κB p65 to 20uL protein binding buffer system, then add 40μL double-stranded DNA probe solution, and incubate at room temperature for 45min. Then add 20 μL Exo III at a concentration of 2.5U / μL, 40 μL MB-1 and 80 μL MB-2, the total volume of the system is 200 μL, the concentration of double-stranded DNA probe in this system is 1 μM, and the concentration of MB-1 is 1 μM ...
Embodiment 2
[0052] Embodiment 2 Sensitivity experiment
[0053] According to the method in Example 1, when detecting NF-κB p65, add NF-κB p65 to 20 uL protein binding buffer system, then add 40 μL double-stranded DNA probe solution, and incubate at room temperature for 45 min. Then add 20 μL 2.5U / μL ExoIII, 40 μL MB-1 and 80 μL MB-2, the concentration of double-stranded DNA probe in this system is 1 μM, the concentration of MB-1 is 1 μM, the concentration of MB-2 is 2 μM, and Incubate at 37°C for 1 h.
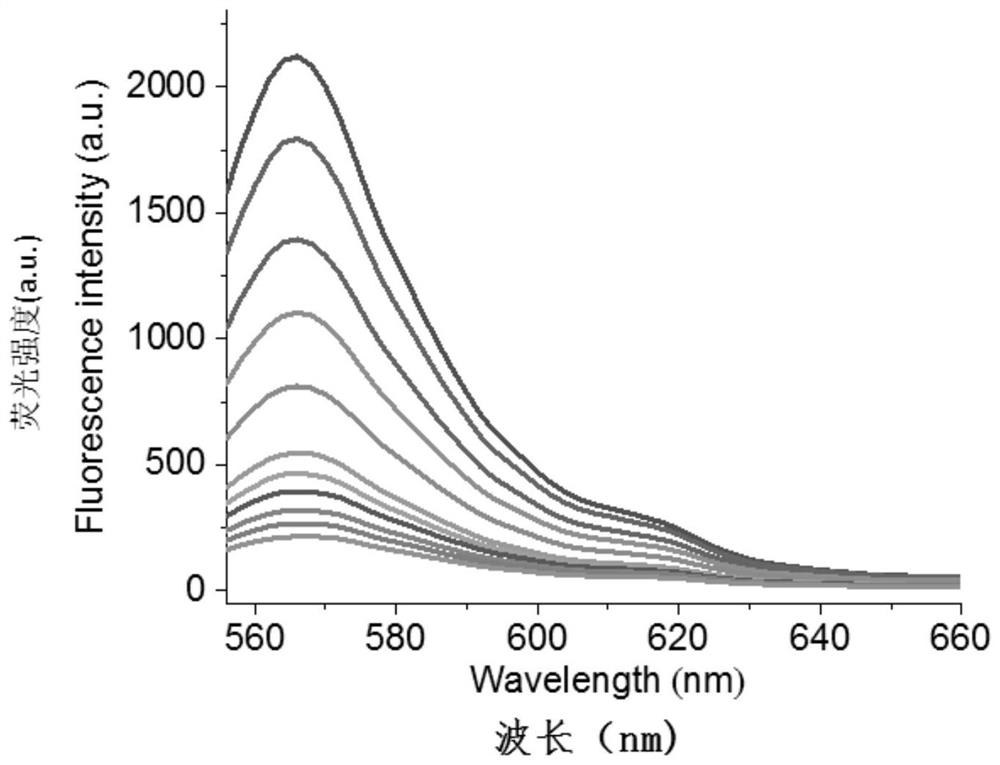
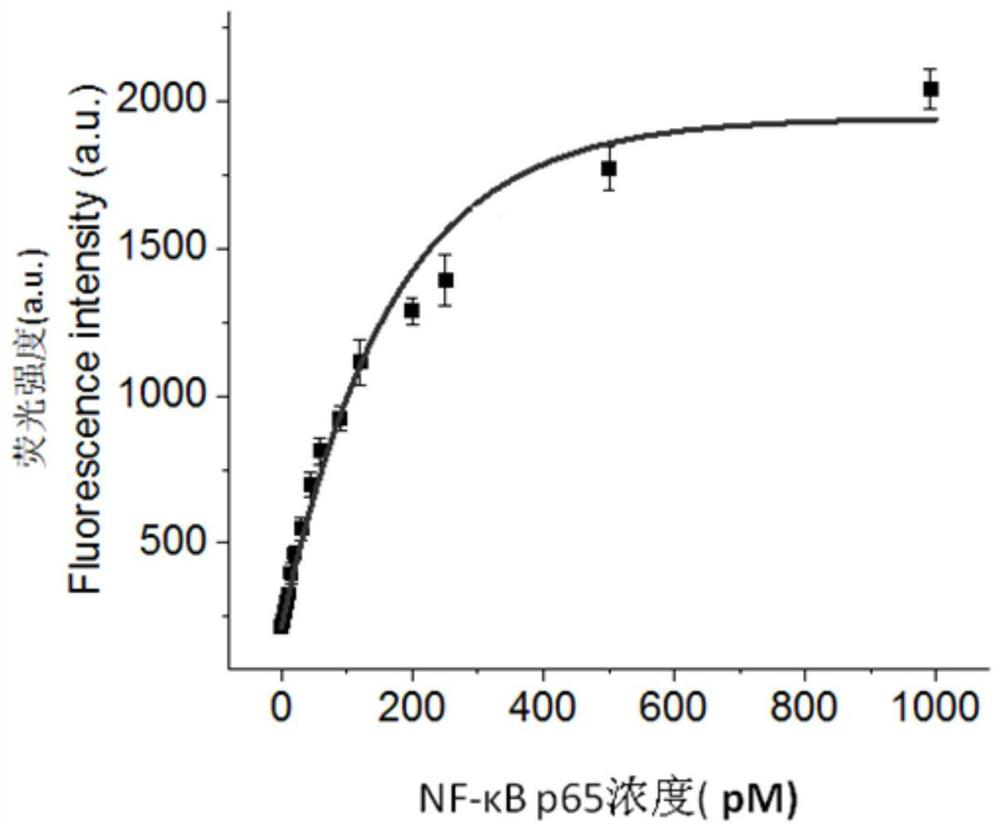
[0054] Detect the fluorescence spectrum of NF-κB p65 at different concentrations (0, 5, 10, 15, 20, 30, 60, 120, 250, 500 and 1000pM) in the range of 556-660nm with a multi-functional microplate reader, the results are shown in figure 2 . From figure 2 It can be seen that the fluorescence signal increases with the increase of NF-κB p65 concentration ( figure 2 The middle curve corresponds to the detection results of NF-κB p65 concentrations of 0, 5, 10, 15, 20, 30, 60, 120, 250, 500...
Embodiment 3
[0055] Embodiment 3 Feasibility study
[0056] Divided into 4 groups:
[0057] Group a: According to the method of Example 1, add 40 μL double-stranded DNA probe solution to 20 μL protein binding buffer system, and incubate at room temperature for 45 minutes; then add 40 μL MB-1 and 80 μL MB-2, and use DNA reaction buffer to complete the system Supplement to 200 μL, the concentration of double-stranded DNA probe in this system is 1 μM, the concentration of MB-1 is 1 μM, and the concentration of MB-2 is 2 μM, incubate at 37°C for 1 hour, and detect the concentration of the system in the range of 556-660nm Fluorescence spectrum; see results in Figure 4 curve a;
[0058] Group b: According to the method of Example 1, NF-κB p65 was added to 20 uL of protein binding buffer system, and then 40 μL of double-stranded DNA probe solution was added, and incubated at room temperature for 45 min. Add 40 μL MB-1 and 80 μL MB-2, and supplement the system to 200 μL with DNA reaction buffe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com