Sporicidal methods and compositions
A composition and spore technology, applied in the direction of organic/inorganic compound composition, botanical equipment and method, biocide, etc., can solve the problems such as difficult to kill endospores
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] microorganism All microorganisms were purchased as spores from certified producers. The microorganisms and manufacturers are listed below.
[0042] Paenibacillus chiba-American Type Culture Collection (ATCC);
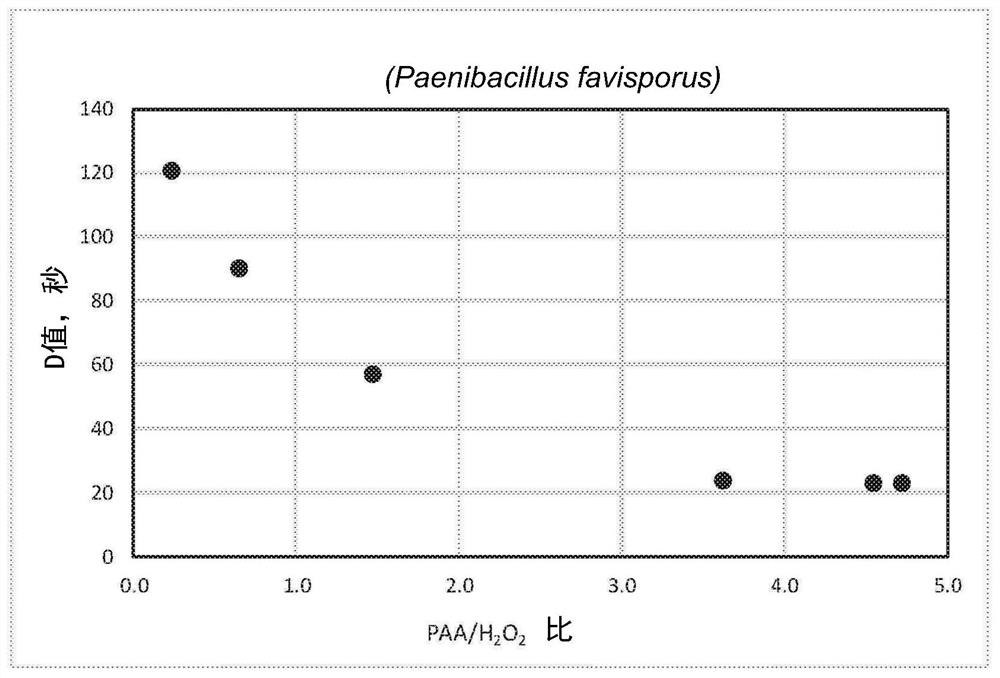
[0043] Paenibacillus favisporus-DSMZ;
[0044] Bacillus cereus 14579 – Presque Isle Cultures;
[0045] Bacillus atrophaeus 9372 – Mesa Labs;
[0046] Bacillus subtilis 6633 – Presque Isle Cultures;
[0047] Bacillus subtilis 19659 – Presque Isle Cultures.
[0048] Upon receipt, by serial dilution in Butterfield buffer and then in 3MAC Petrifilm TM The plate was plated and the titer of the spore culture was determined. Then Petrifilm TM Incubate at 35°C for about 48 hours and count. Store the culture in a dedicated refrigerator. Before each use, re-titre the spore culture to confirm that the culture is still viable and not contaminated.
Embodiment 2
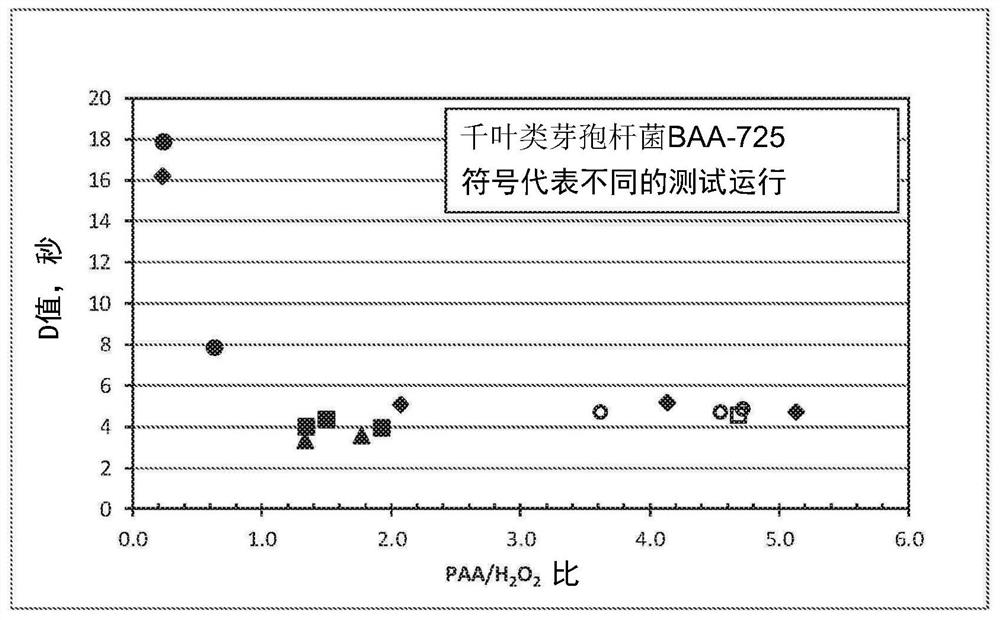
[0050] With PAA / H shown in Table 1 2 o 2 Ratio of PAA formulations to treat Paenibacillus achiba BAA-725.
[0051] Table 1: PAA / H 2 o 2 Preparation
[0052] Preparation 1 2 3 4 5 6 7 Nominal PAA, wt% 5 15 15 22 35 15 22 Nominal H 2 o 2 ,weight%
23 23 10 10 7 3 4 PAA / H 2 o 2 Compare
0.2 0.6 1.5 2.2 5.0 5.0 5.0
[0053] The PAA formulation was diluted to about 2900 ppm in deionized water before use. The temperature of the formulation was raised to 55 °C and the actual PAA and H were measured in an automatic titrator just before use. 2 o 2 concentration.
[0054] Paenibacillus chiliba BAA-725 was inoculated onto stainless steel bars. Exposure to Paenibacillus chiliba BAA-725 was performed by dipping the bar into the PAA formulation for 10 seconds, 20 seconds or 30 seconds. All viable microorganisms were recovered, cultured and counted. Microorganisms were recovered by suspending the culture in Letheen ...
Embodiment 3
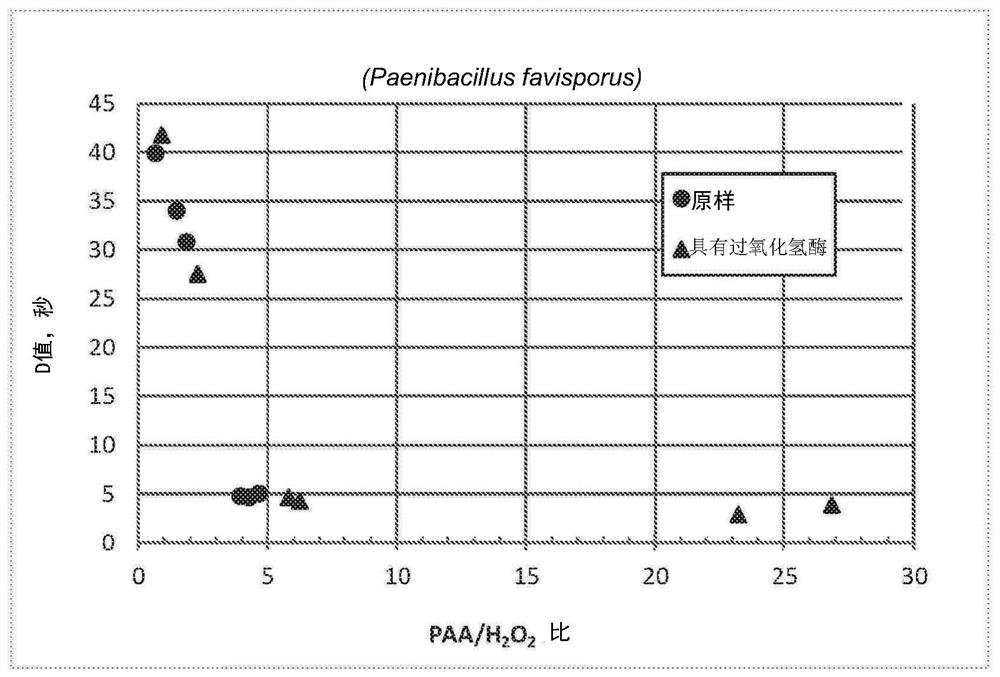
[0058] In the presence and absence of catalase (a hydrolyzed H 2 o 2 In the case of the enzyme), Paenibacillus chiliba BAA-725 was treated with the PAA formulation.
[0059] PAA / H with deionized (DI) water 2 o 2 Equilibrium solution (with 35% by weight of PAA and 7% by weight of H 2 o 2 ) was diluted to a PAA concentration of approximately 2900ppm. For catalase treatment, add 1 μl of catalase (Sigma-Aldrich, catalase from Aspergillus, ≥4000 units / mg protein, about 14.4 mg / ml protein) to 200 ml solution, and Incubate at temperature for 45 minutes. The solution was then warmed to 55°C in a water bath. The heating step took about 38 minutes. After reaching the target temperature, measure the actual PAA and H with an automatic titrator 2 o 2 concentration.
[0060] According to the method described in Example 1, stainless steel bars were inoculated with Paenibacillus chiliba BAA-725 and contacted with catalase-digested PAA solution and non-catalase-digested control PAA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com