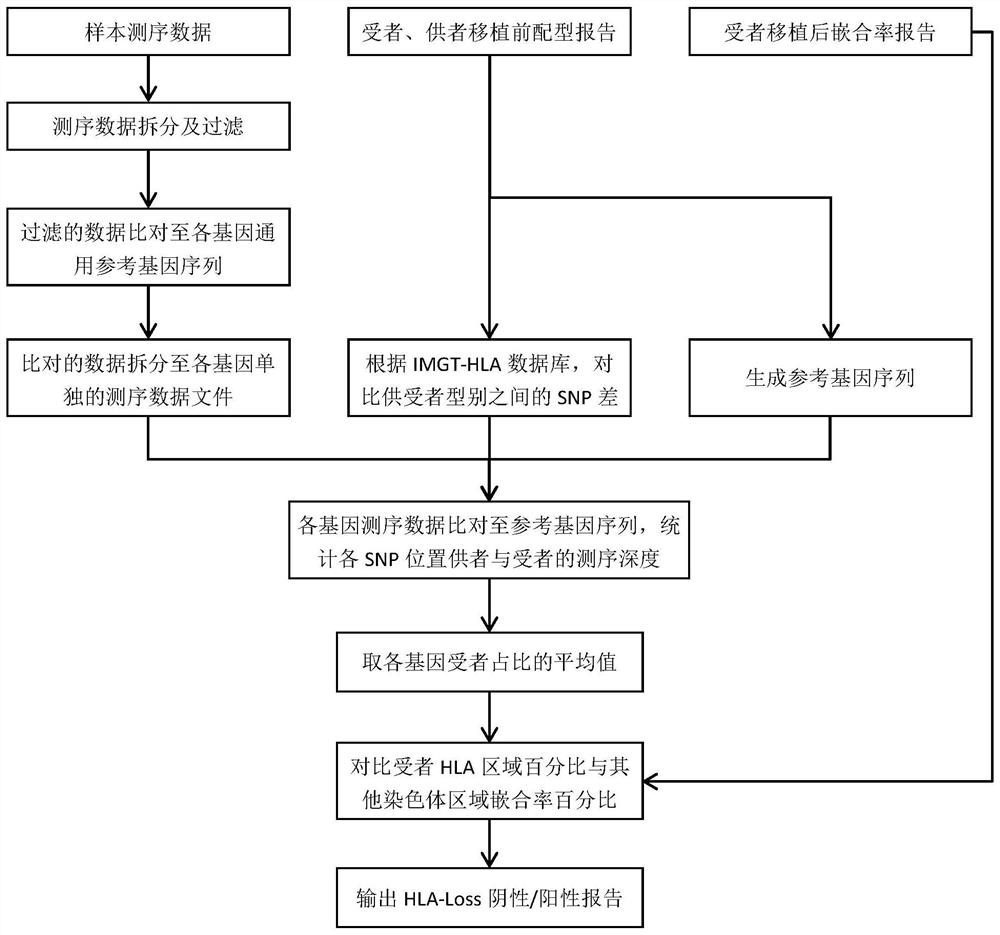
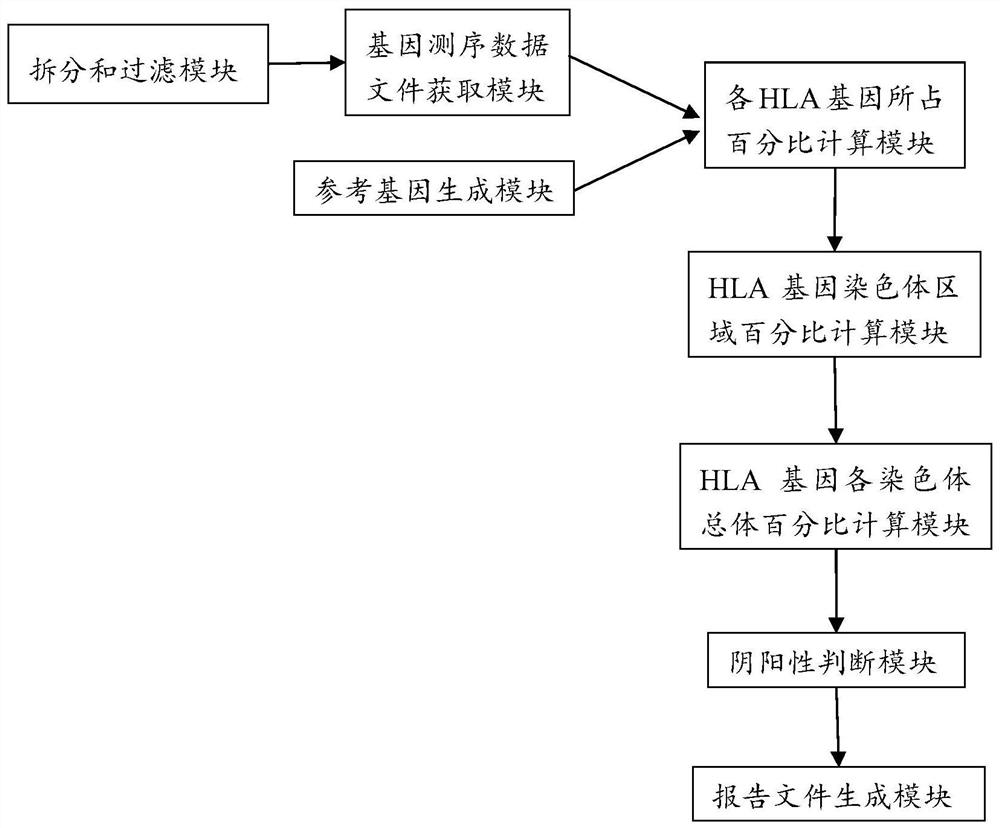
Analysis method and analysis processing device for heterozygous deletion of human HLA chromosome region
A technology for loss of heterozygosity and analysis method, which is applied in the field of analysis method and analysis processing device for loss of heterozygosity in human HLA chromosomal regions, and achieves the effect of small manual interpretation workload, convenient flow and batch operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1: Detection of Heterozygosity Negative Recipients in HLA Chromosomal Regions After Hematopoietic Stem Cell Transplantation
[0057] 1. Perform targeted sequencing testing on hematopoietic stem cell transplant recipients to obtain matching and chimerism results
[0058] In this example, the postoperative blood samples of hematopoietic stem cell transplantation recipients were used, and the genomic DNA was extracted as a template for next-generation targeted sequencing detection, and the detection results were analyzed by the analysis method of the present invention.
[0059] The HLA matching results of the pre-transplant recipients obtained from the pre-transplant matching report are: A*02:10, A*24:02, B*15:01, B*40:01, C*03:03, C*07 :02,DRB1*09:01,DRB1*14:05,DQB1*05:03,DQB1*03:03; donor HLA matching results are: A*24:02,A*26:01,B*15 :01,B*40:01,C*03:03,C*07:02,DRB1*14:05,DRB1*15:02,DQB1*05:03,DQB1*06:01. The B gene and the C gene of the donor and the recipien...
Embodiment 2
[0073] Example 2: Detection of positive recipients with loss of heterozygosity in the HLA chromosomal region after hematopoietic stem cell transplantation
[0074] 1. Perform targeted sequencing detection on recipients after hematopoietic stem cell transplantation, and obtain matching and chimerism results
[0075] In this example, the blood samples of recipients after hematopoietic stem cell transplantation were used, and the genomic DNA was extracted as a template for next-generation targeted sequencing detection, and the detection results were analyzed by the analysis method of the present invention.
[0076] The HLA matching results of the pre-transplant recipients obtained from the pre-transplant matching report are: A*02:03, A*30:01, B*13:02, B*18:01, C*06:02, C*07 :04,DRB1*07:01,DRB1*14:04,DQB1*05:03,DQB1*02:02; donor HLA matching results are: A*11:01,A*30:01,B*13 :02,B*15:02,C*06:02,C*08:01,DRB1*07:01,DRB1*13:02,DQB1*06:04,DQB1*02:02. According to the chimerism rate ...
Embodiment 3
[0089] Embodiment 3: Verify the sensitivity and specificity of the algorithm of the present invention
[0090] In this example, by artificially mixing two healthy human samples in equal proportions, simulating different donor and recipient chimerism rates, using the algorithm of the present invention to detect the proportion of the sequencing results of the two samples at each concentration gradient, and verifying by linear fitting The algorithm sensitivity and specificity of the present invention.
[0091] In this example, two blood samples from healthy people were randomly selected, and the HLA matching results of sample 1 were A*26:01, A*31:01, B*40:06, B*40:06, C*08:01 ,C*15:02,DRB1*09:01,DRB1*15:02,DQB1*06:02,DQB1*03:03, the HLA matching result of sample 2 is A*24:02,A*24:02 ,B*40:01,B*40:03,C*03:04,C*07:02,DRB1*12:01,DRB1*15:01,DQB1*06:02,DQB1*03:01, using After the spectrophotometer detects the DNA concentration of the sample, the two samples are mixed into six differ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com