Gene PeMYB4 sequence for regulating and controlling petal color of phalaenopsis equestris and application thereof
A Phalaenopsis petal and gene sequence technology, which can be applied in application, genetic engineering, plant genetic improvement, etc., can solve the problem of anthocyanin accumulation and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Cloning of embodiment 1 Phalaenopsis PeMYB4 gene
[0036] The total RNA of wild-type Phalaenopsis petals was extracted, the extraction kit was RNAplant (commercially available), and the total RNA was reverse-transcribed into cDNA using a reverse transcription kit (commercially available). Primers were designed according to the transcriptome sequencing results. The primer sequences are shown in SEQ ID NO.3 and SEQ ID NO.4. A 738bp band was amplified from the Phalaenopsis cDNA by RT-PCR. The PCR product was recovered to obtain the PeMYB4 gene, its nucleotide sequence is shown in SEQ ID NO.1, and the amino acid sequence encoded by the nucleotide sequence is shown in SEQ ID NO.2, consisting of 245 amino acid residues, with a molecular weight of 28.35 kilodaltons.
Embodiment 2
[0037] Example 2 Verification of the expression profile of Phalaenopsis phalaenopsis in Xiaolanyu at different developmental stages
[0038] (1) extract as figure 1 As shown, the RNA of Phalaenopsis japonica at different developmental stages was extracted using RNAplant (commercially available), and the total RNA was reverse-transcribed into cDNA using a reverse transcription kit (commercially available).
[0039] (2) Design primers according to transcriptome sequencing data, and the primer sequences are shown in SEQ ID NO.5 and SEQ ID NO.6.
[0040] (3) Using the cDNA obtained by reverse transcription of Phalaenopsis japonica at different developmental stages as a template, the expression profile of PeMYB4 at different developmental stages was verified as follows: figure 2 As shown, the expression of PeMYB4 was significantly induced at different developmental stages. It indicated that PeMYB4 may be involved in the formation of petal color of Phalaenopsis in Xiaolanyu.
Embodiment 3
[0041] Example 3 Verification of the expression profile of PeMYB4 in different tissues of Xiaolanyu Phalaenopsis
[0042] (1) extract as image 3As shown, the RNA of Phalaenopsis xiaolanyu in different tissue parts was extracted using RNAplant (commercially available), and the total RNA was reverse-transcribed into cDNA using a reverse transcription kit (commercially available).
[0043] (2) Design primers according to transcriptome sequencing data, and the primer sequences are shown in SEQ ID NO.5 and SEQ ID NO.6.
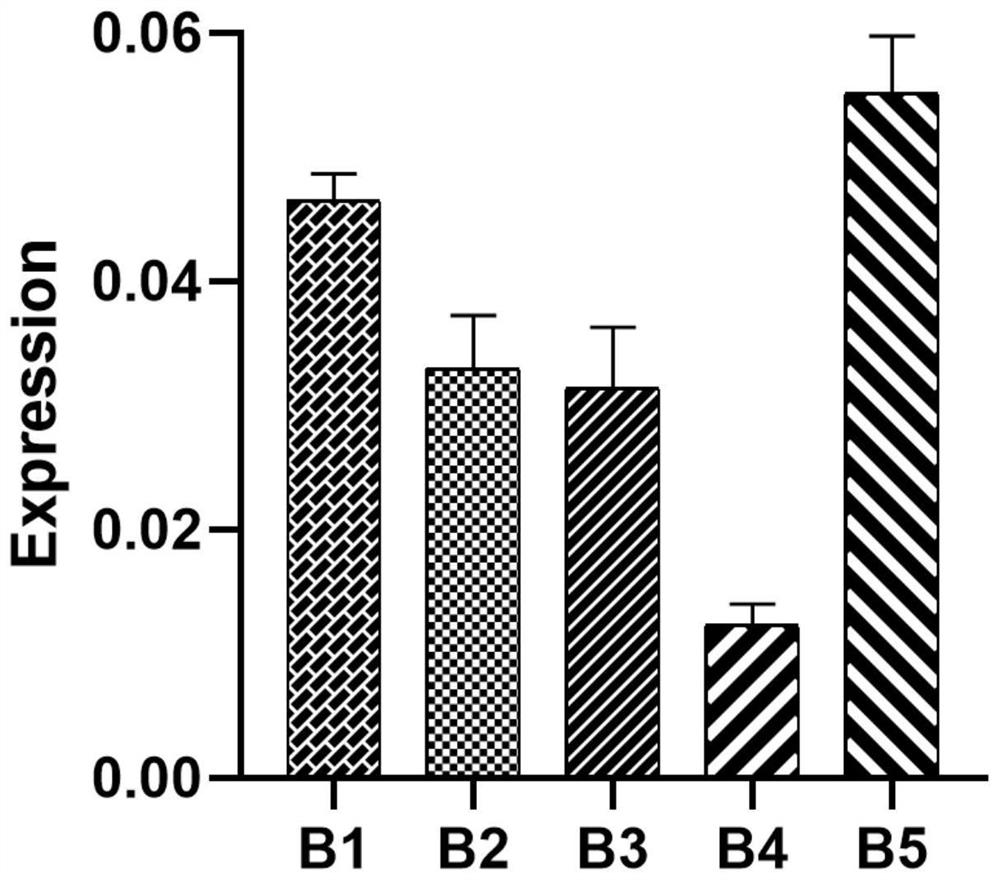
[0044] (3) Using the cDNA obtained by reverse transcription of Phalaenopsis orchid in different tissue parts as a template, the expression profile of PeMYB4 in different tissue parts was verified as follows: Figure 4 As shown, the expression of PeMYB4 is different in different tissues. Higher expression in petals, lips and sepals. It indicated that PeMYB4 may be involved in the formation of petal color of Phalaenopsis orchid in Xiaolanyu.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com